Javier Paz-Ares
Group Leader
Research summary
Plant responses to phosphate starvation represent an emblematic system for studies on regulation of gene activity. In plants, these responses involve both biochemical and developmental changes that improve Pi acquisition and recycling, and protect against the stress of Pi starvation. The induction of Pi starvation responses requires a sophisticated regulatory system that integrates information on external and internal plant Pi concentration, and on other nutrient content. Our aim is to contribute to the dissection of this regulatory system, which operates mainly at the transcriptional level but also involves post-transcriptional control.
Publications
García-León M, Cuyas L, El-Moneim DA, Rodriguez L, Belda-Palazón B, et al. Arabidopsis ALIX regulates stomatal aperture and turnover of abscisic acid receptors. Plant Cell 2019; 10: 2411-2429.
Castrillo G, Teixeira PJ, Paredes SH, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield NW, Mieczkowski P, Jones CD, Paz-Ares J, Dangl JL. Root microbiota drive direct integration of phosphate stress and immunity. Nature 2017; 543: 513-518
Puga MI, Rojas-Triana M, de Lorenzo L, Leyva A, Rubio V, Paz-Ares. J3 Novel signals in the regulation of Pi starvation responses in plants: facts and promises. Curr Opin Plant Biol 2017; 39: 40-49.
Mohan TC, Castrillo G, Navarro C, Zarco-Fernández S, Ramireddy E, Mateo C, Zamarreño AM, Paz-Ares J, Muñoz R, García-Mina JM, Hernández LE, Schmülling T, Leyva A. Cytokinin Determines Thiol-Mediated Arsenic Tolerance and Accumulation. Plant Physiol 2016; 171(2):1418-2.
Cardona-López X, Cuyas L, Marín E, Rajulu C, Irigoyen ML, Gil E, Puga MI, Bligny R, Nussaume L, Geldner N, Paz-Ares J*, Rubio V* (*corresponding authors). ESCRT-III-Associated Protein ALIX mediates High Affinity Phosphate Transporter Trafficking to Maintain Phosphate Homeostasis in Arabidopsis. Plant Cell 2015; 27:2560-81
Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, Miñambres M, Walther D, Schulze WX, Paz-Ares J*, Scheible WR*, Kragler F* (*corresponding authors). Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat Plants 2015; 1(4):15025
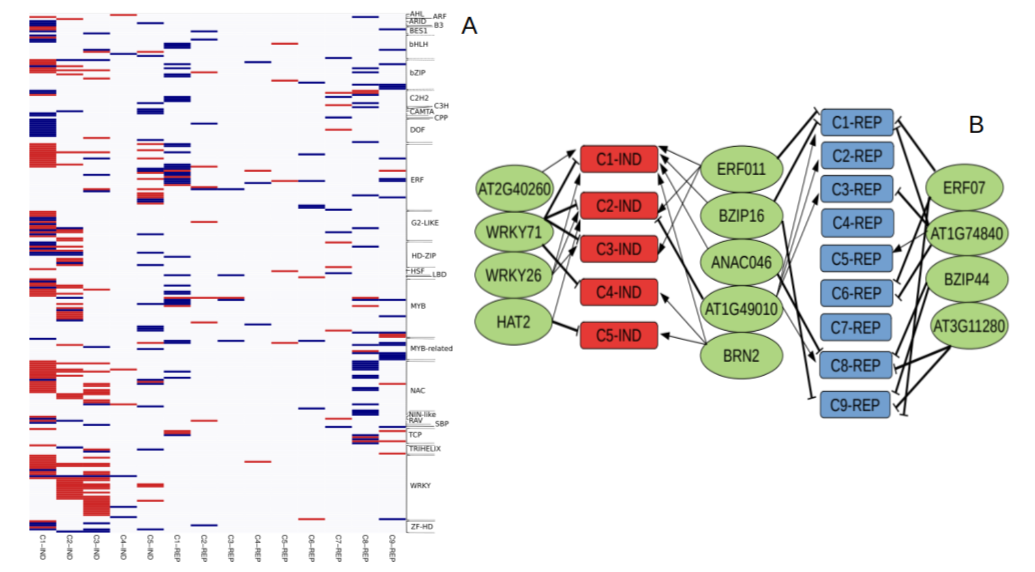
We focus our study on the plant phosphate (Pi) starvation rescue system, which consists of an array of developmental, physiological and molecular responses that allow plants to cope with growth under Pi limiting conditions. This rescue system is a suitable model for studies on regulation of gene activity, and in addition, recently it has attracted considerable interest due to its potential to help design plants with increased Pi acquisition and use efficiency, a necessary requirement to implement low-input sustainable agricultural practices. In the past two years, our main activity has been to exploit natural variation to identify QTLs controlling transcription of Pi starvation genes and affecting Pi acquisition and use efficiency. Our transcriptomic analysis of recombinant inbred lines and natural accessions allowed the identification of a large set of transcription factors controlling expression of Pi starvation responsive genes (Figure 1). And the use of GWAS approaches have uncovered candidate genes affecting growth under Pi limiting conditions (Figure 2), whose characterisation is underway.
Additionally, we have examined the dynamics of interchromatin interactions in response to Pi starvation using Hi-C related approaches. We found no large effects of Pi starvation on chromatin interactions, but observed that genes induced by Pi starvation (PSI) tend to display increased chromatin interconnections among themselves, indicating a constitutive predisposition for coordinated PSI gene expression
Finally, we have also initiated an study of extrachromosomal circular DNA formation in response to Pi starvation. It is presently well established that eccDNA formation is a widespread characteristic of eukaryotes, where eccDNAs are originated from thousands of locations of their genomes. We have examined eccDNA formation during Pi starvation and in line with expectations more than 1500 eccDNA have been identified, out of which a 3% appear to be Pi starvation specific. We are presently studying their biogenesis and their potential functional significance.








