Urtzi Garaigorta
Group Leader
Research summary
Our laboratory is interested in unraveling the nature of the virus-host interactions and pathways that regulate the formation and homeostasis of the hepatitis B virus cccDNA pool, a hallmark of virus persistence. We combine genetic screenings with virological, cellular and molecular approaches to identify and characterize the mode-of-action of new cellular factors that regulate the HBV infection.
Publications
I. Galindo, U. Garaigorta, F. Lasala, M.A. Cuesta-Geijo, P. Bueno, C. Gil, R. Delgado, P. Gastaminza, C. Alonso. Antiviral drugs targeting endosomal membrane proteins inhibit distant animal and human pathogenic viruses. Antiviral Res., 2020, 26:104990.
L. Marcos-Villar, E. Nistal-Villan, N. Zamarreño, U. Garaigorta, P. Gastaminza, A. Nieto. Interferon-β Stimulation Elicited by the Influenza Virus Is Regulated by the Histone Methylase Dot1L through the RIG-I-TRIM25 Signaling Axisβ Stimulation Elicited by the Influenza Virus Is Regulated by the Histone Methylase Dot1L through the RIG-I-TRIM25 Signaling Axis. Cells, 2020, 9(3):732.
C. Whitten-Bauer, J. Chung, A. Gómez-Moreno, P. Gomollón-Zueco, M.D. Huber, L. Gerace, U. Garaigorta. The Host Factor Erlin-1 is Required for Efficient Hepatitis C Virus Infection. Cells, 2019, 8(12):1555.
Billioud G, Kruse RL, Carrillo M, Whitten-Bauer C, Gao D, Kim A, Chen L, McCaleb ML, Crosby JR, Hamatake R, Hong Z, Garaigorta U, Swayze E, Bissig KD, Wieland S. In vivo reduction of hepatitis B virus antigenemia and viremia by antisense oligonucleotides. Journal of Hepatology 2016; 64 (4):781-789.
Padmanabhan P, Garaigorta U, Dixit MD. Emergent properties of the interferon signaling network may underlie the success of hepatitis C treatment. Nature Communications 2014; 16 (5):3872-3880.
Funding
 2016-2019: Convocatoria 2016 - Proyectos I+D+I. Programa estatal de investigación, desarrollo e innovación orientada a los retos de la sociedad. Regulation of hepatitis B virus infection by DNA damage response proteins. Ref. SAF2016-75169-R.
2016-2019: Convocatoria 2016 - Proyectos I+D+I. Programa estatal de investigación, desarrollo e innovación orientada a los retos de la sociedad. Regulation of hepatitis B virus infection by DNA damage response proteins. Ref. SAF2016-75169-R.
Our laboratory is interested in understanding virus host interactions that regulate the outcome and pathogenesis of virus infections. Our main objective is to identify vulnerabilities that could be exploited to develop new antiviral therapies. In the last years we have used hepatitis B virus (HBV) and hepatitis C virus (HCV) infection cell culture models. These hepatic viruses are responsible of millions of cases of acute and chronic hepatitis and represent the major etiological agent of liver cancer worldwide.
During the 2019-2020 period, we focused on understanding the role of cellular proteins in the virus life cycle. On one hand, we identified Erlin-1 protein, an endoplasmic reticulum resident protein, as a new host factor required for efficient HCV infection. Gene silencing experiments have demonstrated that Erlin-1 protein regulates early as well as late steps in the HCV life cycle. Interestingly, Erlin-2, a protein with high sequence and functional homology with Erlin-1 protein does not play any important role in HCV infection. Our results provide new insights into functional differences between the two Erlins and identify a new molecular target for therapeutic intervention. On the other hand, we have confirmed and expanded our initial observations that DNA damage response related proteins are key restriction factors for HBV infection. Moreover, we are working on basic aspects of HBV DNA integration, key for cancer development.
Since the SARS-CoV-2 pandemic started our group have teamed up with Dr. Gastaminza’s group to establish the CNB Antiviral Platform. The main objective is the identification and characterization of new antiviral compounds against highly pathogenic human virus infections. We have screened thousands of chemical compounds and identified new families of experimental compounds with antiviral activity against SARS-CoV-2. Moreover, we have identified repurposing drugs that are been considered for clinical testing. Finally, we have signed several research contracts with the pharmaceutical industry.
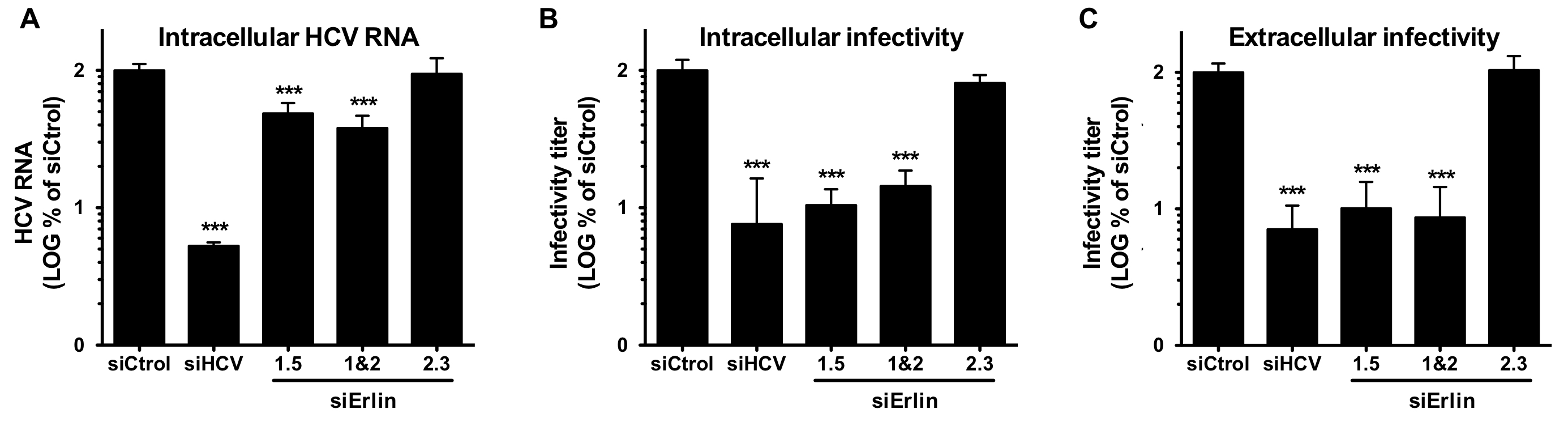
Erlin-1 protein down-regulation interferes with HCV infection, shown by reduced levels of intracellular RNA (A) and infectivity (B) as well as extracellular infectivity (C).
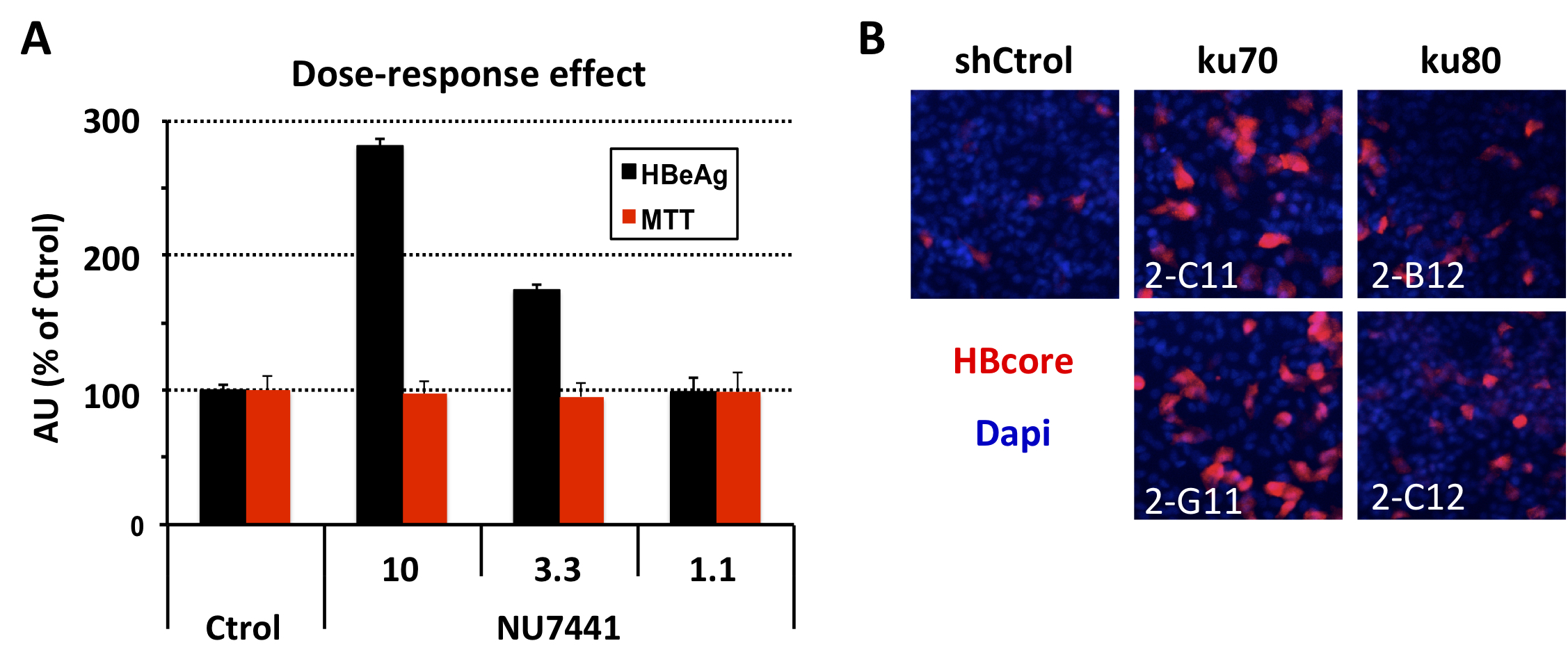
(A) Treatment with the DNA-PKc inhibitor NU7441 increases HBeAg levels in a de novo HBV infection (B) Silencing of ku70 and ku80 proteins, regulator subunits of DNA-PKc, increases the intracellular accumulation of HBV core protein.








