Juan Antonio García
Group Leader
Carmen Simón
Group Leader
Research summary
Plant viruses depend largely on host factors to replicate in the cell and to propagate throughout the plant and between individual plants. Plants in turn have developed antiviral defence mechanisms that must be counteracted by viral factors. These factors appear to be preferred targets for alternative plant defences. In our laboratory, we try to understand this complex interplay, mainly in the infection of the potyvirus Plum pox virus (PPV), the causal agent of sharka, a damaging disease of Prunus trees. We are especially interested in defence responses related to RNA silencing and its viral suppressors.
Publications
González de Prádena A, Sánchez Jiménez A, San León D, Simmonds P, García JA, Valli AA. Plant virus genome is shaped by specific dinucleotide restrictions that influence viral infection. MBio 2020; 11: e02818-02819
Hervás M, Navajas R, Chagoyen M, Garcia JA, Martinez-Turiño, S. Phosphorylation-related cross-talk between distant regions of the core region of the coat protein contributes to virion assembly of Plum pox virus. Mol Plant Microbe Interact 2020; 33: 653-667
Hervás M, Ciordia S, Navajas R, García JA, Martínez-Turiño S. Common and strain-specific post-translational modifications of the potyvirus Plum pox virus coat protein in different hosts. Viruses 2020; 12: 308.
Pasin F, Shan H, García B, Müller M, San León D, Ludman M, et al. Abscisic acid connects phytohormone signaling with RNA metabolic pathways and promotes an antiviral response that Is evaded by a self-controlled RNA virus. Plant Commun 2020; 1: 100099.
Ochoa J, Valli A, Martín-Trillo M, Simón-Mateo C, García JA, Rodamilans B. Sterol isomerase HYDRA1 interacts with RNA silencing suppressor P1b and restricts potyviral infection. Plant Cell Environ 2019; 42: 3015-3026
Gallo A, Valli A, Calvo M, García JA. A functional link between RNA replication and virion assembly in the potyvirus Plum pox virus. J. Virol. 2018; 92, e02179-17.
Plants are frequently infected in nature by viruses. Most of these infections are symptomless, or even give rise to mutualist associations, but plant viruses can also cause severe diseases. Breeding for resistance has been useful to fight some viral diseases, however, natural sources of resistance are scarce. The development of genetic engineering has expanded the available arsenal to generate virus-resistant plants. Understanding natural resistance mechanisms and viral amplification processes is essential to find appropriate targets for biotechnological antiviral strategies.
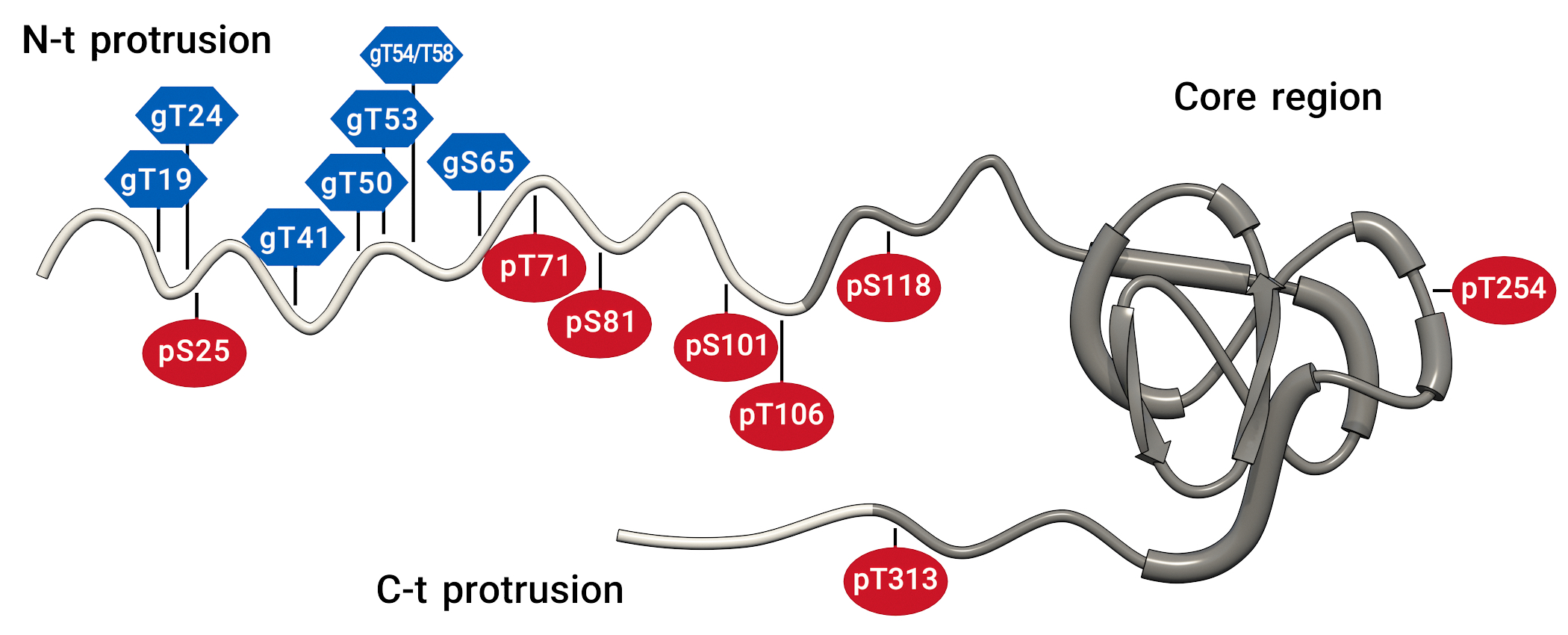
Our research aims to contribute to meet this need. We are mainly interested in the family Potyviridae, especially in Plum pox virus, which causes sharka, a devasting disease of trees of the genus Prunus. In these two years we have paid attention to two viral functions that still have not been intensively studied, the proteolytic processing of viral polyproteins and the post-translational modifications (PTMs) of viral proteins. We have shown that the efficiency of the potyviral leader protease may be restricted to avoid that the uncontrolled release of the silencing suppressor HCpro triggers antiviral defences through complex hormonal and transcriptomic changes. We have also obtained data suggesting that alteration of the proteolytic cleavage between NIapro and VPg proteins is involved in the unique known escape of PPV from the HR-like resistance of some Prunus domestica cultivars. Regarding PTMs, our results have led us to propose that, whereas joint and opposite action of O-GlcNAcylation and phosphorylation at the N-terminal protrusion of the PPV capsid protein regulates the stability of this factor, phosphorylation at its core region controls assembly and disassembly of viral particles. Other remarkable results have been the finding that the sterol isomerase HYDRA1 restricts PPV infection and the demonstration that the viral genomic sequence is shaped by specific dinucleotide restrictions, so that an increase in UpA frequency causes a strong reduction of virus accumulation. [Research supported by grants of the Spanish government BIO2016-80572-R and PID2019-109380RB-100 (IPs J.A. García and Carmen Simón) BIO2017-92613-EXP (IP C. Simón), and BIO2015-73900-JIN and PID2019-110979RB-100 (IP A. Valli)]

Figure legend. Unraveling the mechanism of induction of hypersensitive response associated to resistance to Plum pox virus in European plums. (a) Resistant Prunus domestica trees inoculated with standard PPV-D isolate, the resistance-escaping isolate PPV-DH or a chimeric virus carrying the PPV-D background with the NIa sequence of PPV-DH. Schematic representation of each virus can be seen on the bottom of each picture. (b) In planta expression of NIa proteins from PPV-D and PPV-DH with a Myc tag to detect cleavage activity by western blot. Quantification of the cleavage percentage observed for each protease is shown in the panel.
 Laboratory Members
Laboratory Members








