José R. Castón
Group Leader
Research summary
Our studies address to elucidate structure-function relationships of viral macromolecular complexes, also known as viral nanomachines, which control many fundamental processes in virus life cycle. For that, we have incorporated state-of-the-art approaches to obtaining near-atomic resolution structure directly from electron microscope images..
Publications
Rodríguez-Espinosa MJ, Rodríguez JM, Castón JR, de Pablo PJ. Mechanical disassembly of human picobirnavirus like particles indicates that cargo retention is tuned by the RNA-coat protein interaction. Nanoscale Horizons 2023; 8: 1665-1676.
Luque D, Ortega-Esteban A, Valbuena A, Vilas JL, Rodríguez-Huete A, Mateu MG, Castón JR. Equilibrium dynamics of a biomolecular complex analyzed at single-amino acid resolution by cryo-electron microscopy. J Mol Biol 2023; 435: 168024.
Castón JR, Zlotnick A. Virus structure and expression. Curr Opin Virol 2022, 52, 101277
Luque D, Goulas T, Mata CP, Mendes SR, Gomis-Rüth FX, Castón JR. Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human α2-macroglobulin. Proc Natl Acad Sci USA 2022; 119 (19): e2200102119.
Luque D, Castón JR. Cryo-electron microscopy for the study of virus assembly. Nat Chem Biol. 2020;231-239.
 Our studies address to elucidate structure-function-evolution relationships of viral macromolecular complexes, also known as viral nanomachines, which control many fundamental processes in virus life cycle. For that, we have incorporated state-of-the-art approaches to obtaining high-resolution structures of viral assemblies in near-native conditions, at resolutions better than 3 Å by combining data from several hundreds or often thousands of electron microscope images.
Our studies address to elucidate structure-function-evolution relationships of viral macromolecular complexes, also known as viral nanomachines, which control many fundamental processes in virus life cycle. For that, we have incorporated state-of-the-art approaches to obtaining high-resolution structures of viral assemblies in near-native conditions, at resolutions better than 3 Å by combining data from several hundreds or often thousands of electron microscope images.
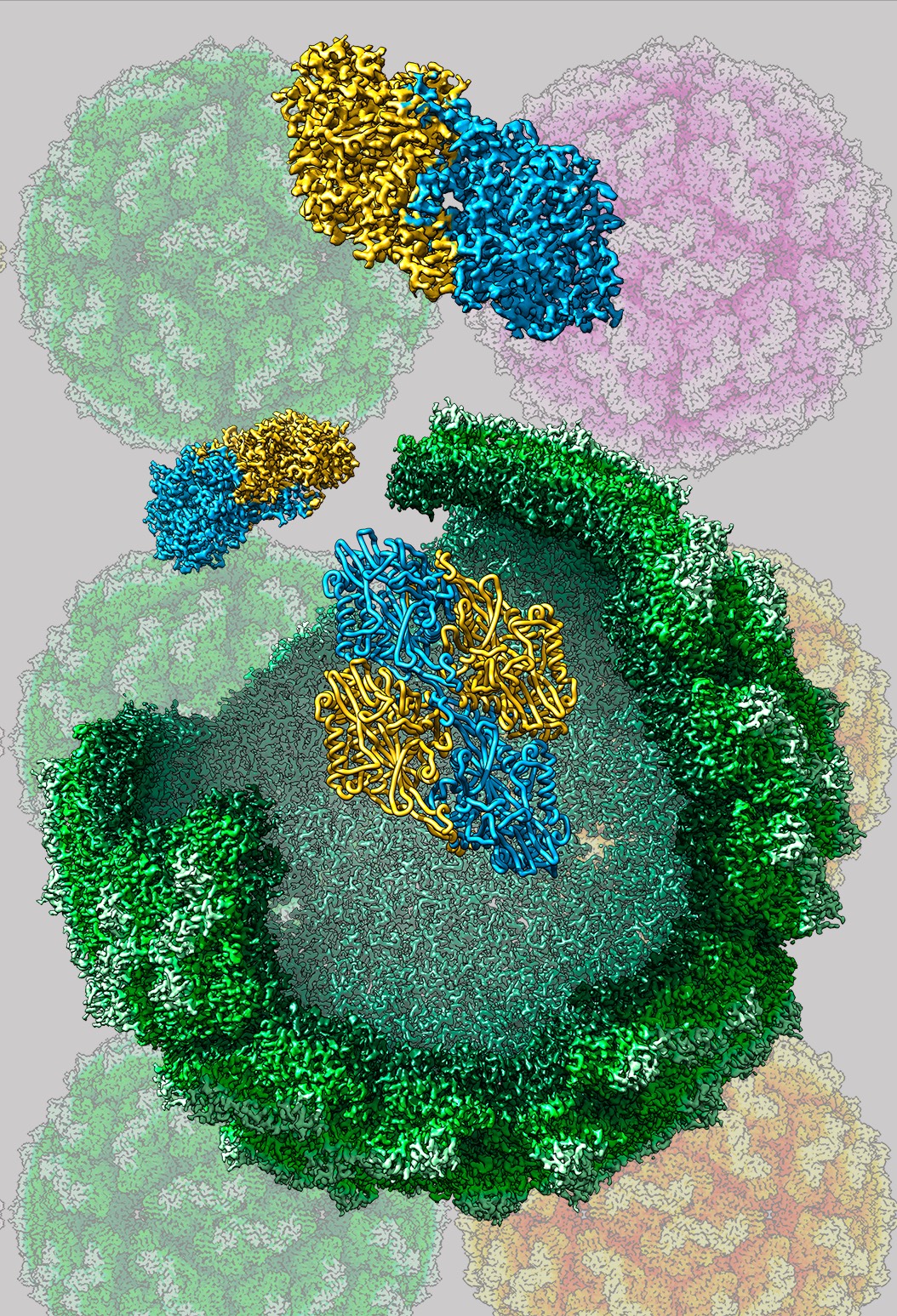
Capsids are dynamic structures whose components have transient conformations associated with specific roles in the viral cycle. In addition, the capsid is a metastable assembly: it is robust enough to protect the genome, and labile enough to allow genome delivery into the host cell. Information on virus structures at the highest possible resolution is essential for identifying their molecular mechanisms and functions. Three-dimensional cryogenic electron microscopy (3D cryo-EM) of viruses and viral capsids is widely used to determine their structures at near-atomic resolution in near-native conditions. Structural analysis of viruses is complemented by study of mechanical properties by atomic force microscopy (AFM), to examine the relationship between physical properties such as rigidity and mechanical resilience, and virus biological function. Our basic structural research shows alternatives for interfering in their function, as well as clues for vaccine design and/or antiviral drugs. These studies are also central to establishing the basis for incorporation of heterologous proteins, nucleic acids, and/or chemicals into viral capsids (considered as nanocontainers), of potential use for future biotechnological applications.
Our group studies the structure and conformational polymorphism of several viral systems, including double-stranded RNA (dsRNA) viruses such as birnaviruses (infectious bursal disease virus, IBDV), human picobirnavirus (HPBV) and several fungal viruses, as well as single-stranded RNA viruses such as rabbit hemorrhagic disease virus (RHDV) and human rhinovirus (HRV). In collaboration with several national and international groups, we extend our studies to other bacterial and eukaryotic complexes such as encapsulin and α 2-macroglobulin,respectively.
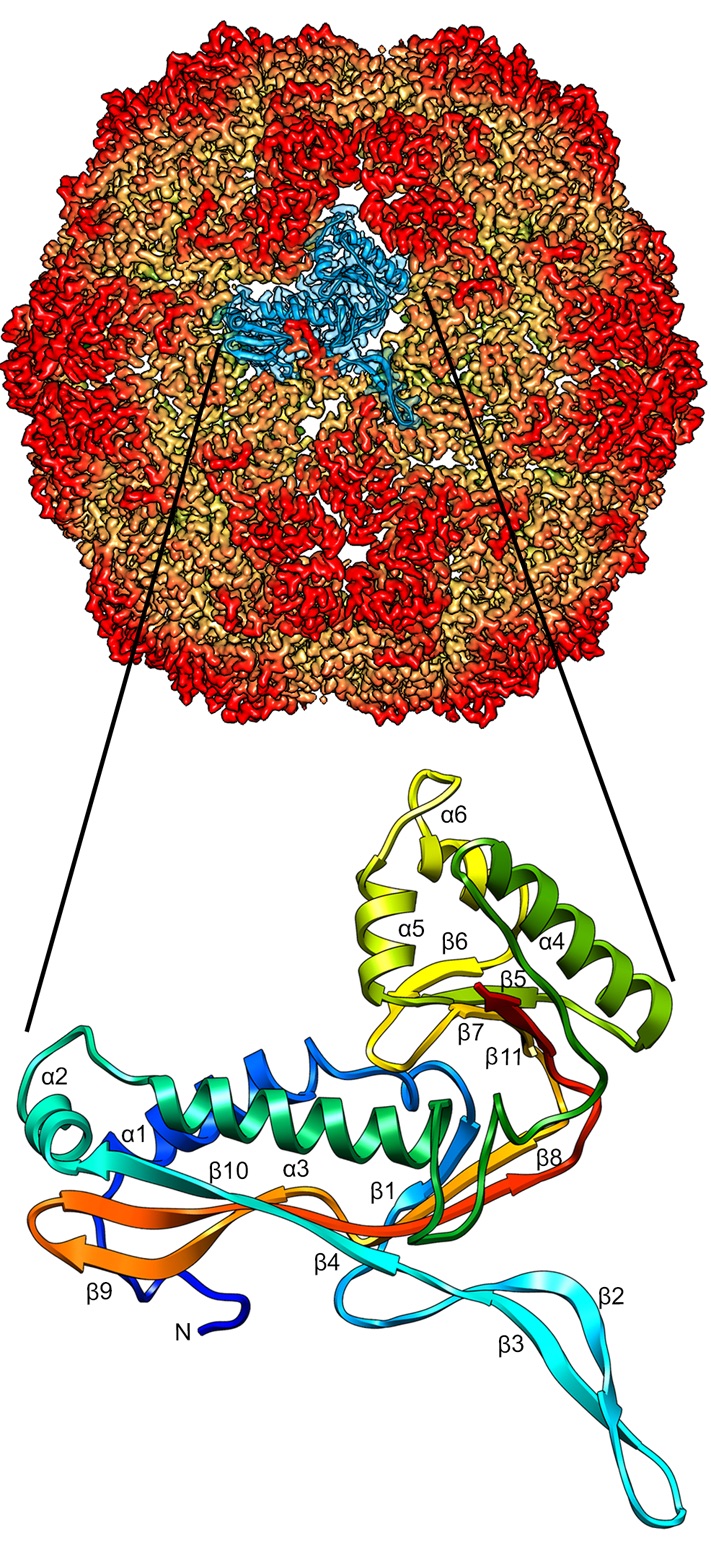
Cryo-EM Structure of Brevibacterium linens encapsulin (BlEnc) at 2.28 Å resolution. Structural comparison between BlEnc and the capsid proteins of the viral lineage HK97 shows high structural similarity, indicating that both compartments descend from a common ancestor.








