Luis Ángel Fernández
Group Leader
Research summary
Our work focuses on engineering E. coli bacteria for biomedical applications, including the selection and production of small recombinant antibodies in bacteria and the design of bacteria for use in diagnosis and therapy in vivo. We study protein secretion systems found in pathogenic E. coli strains and modified them to become nanomachines that may be useful in the selection and expression of proteins of therapeutic interest in nonpathogenic strains of E. coli.
Publications
Asensio-Calavia A, Ceballos-Munuera Á, Méndez-Pérez A, Álvarez B, LA. Fernández. A tuneable genetic switch for tight control of tac promoters in Escherichia coli boosts expression of synthetic injectisomes. Microbial Biotechnology 2023, 00, 1–14, DOI: 10.1111/1751-7915.1432
Cerdán L, Álvarez B, LA Fernández. Massive integration of large gene libraries in the chromosome of Escherichia coli. Microbial Biotechnology, 2023; 00, 1–16 . DOI: 10.1111/1751-7915.14367
Casasnovas JM, Margolles Y, Noriega MA, Guzmán M, Arranz R, Melero R, Casanova M, Corbera JA, Jiménez de Oya, Gastaminza P, Garaigorta U, Saiz JC, Martín-Acebes MA, and LA Fernández. Nanobodies protecting from lethal SARS-CoV-2 infection target receptor binding epitopes preserved in virus variants other than omicron. Frontiers in Immunology 2022, 13:863831, DOI: 10.3389/fimmu.2022.863831
Seco EM, LA Fernández. Efficient markerless integration of genes in the chromosome of probiotic E. coli Nissle 1917 by bacterial conjugation. Microbial Biotechnology 2022 15(5):1374-1391.DOI: 10.1111/1751-7915.13967
Álvarez B, Mencía M, de Lorenzo M & Fernández LA In vivo diversification of target genomic sites using processive base deaminase fusions blocked by dCas9. Nat Comms 2020; 11: 6436 https://doi.org/10.1038/s41467-020-20230-z
Ruano-Gallego D, Yara DA, Di Ianni L, Frankel G, Schüller S, Fernández LÁ. A nanobody targeting the translocated intimin receptor inhibits the attachment of enterohemorrhagic E. coli to human colonic mucosa.PLoS Pathog. 2019; 15(8):e1008031. PMID: 31465434 DOI:10.1371/journal.ppat.1008031
 Our research is aimed to engineer E. coli bacteria for biomedical applications, including the selection of small recombinant antibodies and the design of bacteria for diagnostic and therapeutic use in vivo. We study protein secretion systems found in pathogenic E. coli strains and engineer them to develop protein nanomachines that can be applied for selection of recombinant antibodies and the delivery of therapeutic proteins by non-pathogenic E. coli strains. Among the recombinant antibodies, we employ single-domain antibodies (sdAbs) or nanobodies, the smallest antibody fragments known-to-date with full antigen-binding capacity. Nanobodies are based on VHH domains obtained from heavy-chain-only antibodies found in camelids (e.g. dromedaries, llamas). We use synthetic biology approaches and genome engineering to combine the expression of these modular parts in the designed bacteria.
Our research is aimed to engineer E. coli bacteria for biomedical applications, including the selection of small recombinant antibodies and the design of bacteria for diagnostic and therapeutic use in vivo. We study protein secretion systems found in pathogenic E. coli strains and engineer them to develop protein nanomachines that can be applied for selection of recombinant antibodies and the delivery of therapeutic proteins by non-pathogenic E. coli strains. Among the recombinant antibodies, we employ single-domain antibodies (sdAbs) or nanobodies, the smallest antibody fragments known-to-date with full antigen-binding capacity. Nanobodies are based on VHH domains obtained from heavy-chain-only antibodies found in camelids (e.g. dromedaries, llamas). We use synthetic biology approaches and genome engineering to combine the expression of these modular parts in the designed bacteria.
Current projects:
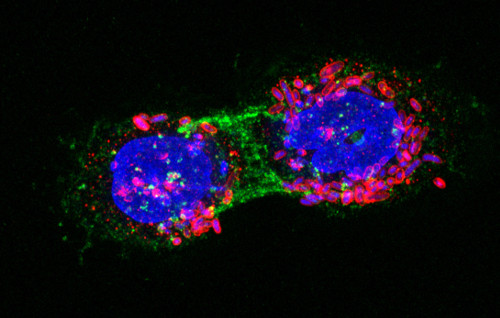
1) Expression and selection of nanobodies against pathogens and cancer. We have used E. coli surface display and protein secretion systems to screen immune libraries of nanobodies and to select high-affinity clones that, for instance, inhibit the adhesion of enterohemorrhagic E. coli (EHEC) to human intestinal cells. We have also selected nanobodies binding relevant antigens in cancer (e.g., EGFR). Our ongoing projects include additional relevant tumor antigens (PD-L1) and neutralizing nanobodies against SARS-CoV-2.
2) Engineering E. coli bacteria as anti-tumor agents. We have continued our synthetic biology project to modify non-pathogenic E. coli chassis with synthetic adhesins and a type III protein secretion system (T3SS) to obtain bacteria with specific anti-tumor activities.
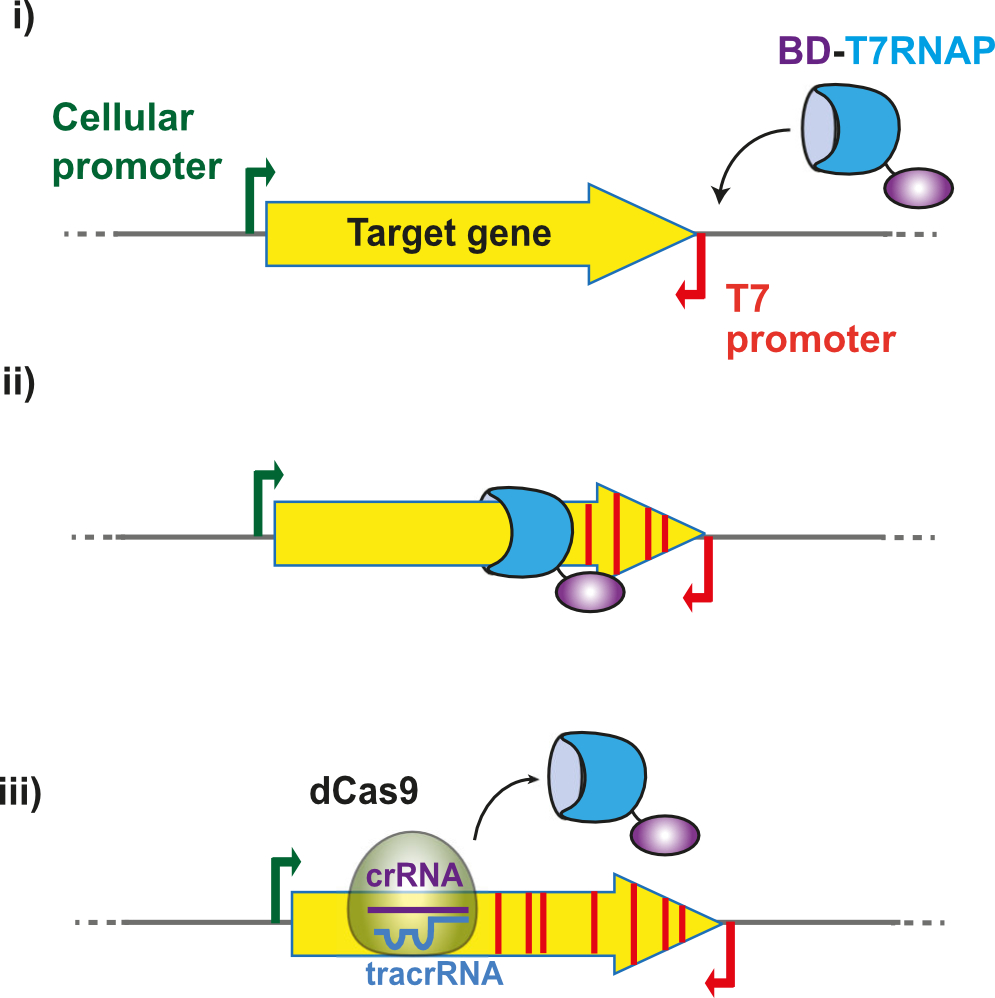
3) Accelerating protein evolution in vivo. We have developed a novel in vivo mutagenesis system in E. coli, called T7-DIVA, based on the recruitment of base deaminases to a target gene with T7 RNA polymerase. This system enables us to accelerate the directed evolution of proteins of interest, such as enzymes and antibodies.