Environ Microbiol Rep. 2013 Apr;5(2):291-300
Sevilla E, Silva-Jiménez H, Duque E, Krell T, Rojo F.
 Sensor kinases play a key role in sensing and responding to environmental and physiological signals in bacteria. In this study we characterized a previously unknown orphan hybrid sensor kinase from Pseudomonas putida, which is conserved in several Pseudomonads.
Sensor kinases play a key role in sensing and responding to environmental and physiological signals in bacteria. In this study we characterized a previously unknown orphan hybrid sensor kinase from Pseudomonas putida, which is conserved in several Pseudomonads.
Inactivation of the gene coding for this sensor kinase, which we have named HskA, modified the expression of at least 85 genes in cells growing in a complete medium. HskA showed a strong influence on the composition of the electron transport chain. In cells growing exponentially in a complete medium, the absence of HskA led to a significant reduction in the expression of the genes coding for the bc1 complex and for the CIO and Cbb3-1 terminal oxidases. In stationary phase cells, however, lack of HskA caused a higher expression of the Cyo terminal oxidase and a lower expression of the Aa3 terminal oxidase. The HskA polypeptide shows two PAS (signal-sensing) domains, a transmitter domain containing the invariant phosphorylatable histidine and an ATP binding site, and a receiver domain containing the conserved aspartate capable of transphosphorylation, but lacks an Hpt module. It is therefore a hybrid sensor kinase. Phosphorylation assays showed that purified HskA undergoes autophosphorylation in the presence of ATP.
Sauer M, Delgadillo MO, Zouhar J, Reynolds GD, Pennington JG, Jiang L, Liljegren SJ, Stierhof YD, De Jaeger G, Otegui MS, Bednarek SY, Rojo E.
 Many soluble proteins transit through the trans-Golgi network (TGN) and the prevacuolar compartment (PVC) en route to the vacuole, but our mechanistic understanding of this vectorial trafficking step in plants is limited. In particular, it is unknown whether clathrin-coated vesicles (CCVs) participate in this transport step.
Many soluble proteins transit through the trans-Golgi network (TGN) and the prevacuolar compartment (PVC) en route to the vacuole, but our mechanistic understanding of this vectorial trafficking step in plants is limited. In particular, it is unknown whether clathrin-coated vesicles (CCVs) participate in this transport step.
Through a screen for modified transport to the vacuole (mtv) mutants that secrete the vacuolar protein VAC2, we identified MTV1, which encodes an EPSIN N-TERMINAL HOMOLOGY protein, and MTV4, which encodes the ADP ribosylation factor GTPase-activating protein NEVERSHED/AGD5. MTV1 and NEV/AGD5 have overlapping expression patterns and interact genetically to transport vacuolar cargo and promote plant growth, but they have no apparent roles in protein secretion or endocytosis. MTV1 and NEV/AGD5 colocalize with clathrin at the TGN and are incorporated into CCVs. Importantly, mtv1 nev/agd5 double mutants show altered subcellular distribution of CCV cargo exported from the TGN. Moreover, MTV1 binds clathrin in vitro, and NEV/AGD5 associates in vivo with clathrin, directly linking these proteins to CCV formation.
These results indicate that MTV1 and NEV/AGD5 are key effectors for CCV-mediated trafficking of vacuolar proteins from the TGN to the PVC in plants.
PLoS One. 2013 Jun 20;8(6):e65986
Goñi-Moreno A, Amos M, de la Cruz F.
Recent efforts in synthetic biology have focussed on the implementation of logical functions within living cells. One aim is to facilitate both internal "re-programming" and external control of cells, with potential applications in a wide range of domains. However, fundamental limitations on the degree to which single cells may be re-engineered have led to a growth of interest in multicellular systems, in which a "computation" is distributed over a number of different cell types, in a manner analogous to modern computer networks. Within this model, individual cell type perform specific sub-tasks, the results of which are then communicated to other cell types for further processing. The manner in which outputs are communicated is therefore of great significance to the overall success of such a scheme. Previous experiments in distributed cellular computation have used global communication schemes, such as quorum sensing (QS), to implement the "wiring" between cell types. While useful, this method lacks specificity, and limits the amount of information that may be transferred at any one time. We propose an alternative scheme, based on specific cell-cell conjugation. This mechanism allows for the direct transfer of genetic information between bacteria, via circular DNA strands known as plasmids.
We design a multi-cellular population that is able to compute, in a distributed fashion, a Boolean XOR function. Through this, we describe a general scheme for distributed logic that works by mixing different strains in a single population; this constitutes an important advantage of our novel approach. Importantly, the amount of genetic information exchanged through conjugation is significantly higher than the amount possible through QS-based communication. We provide full computational modelling and simulation results, using deterministic, stochastic and spatially-explicit methods. These simulations explore the behaviour of one possible conjugation-wired cellular computing system under different conditions, and provide baseline information for future laboratory implementations.
Immunity. 2013 Jun 27;38(6):1176-86
Del Fresno C, Soulat D, Roth S, Blazek K, Udalova I, Sancho D, Ruland J, Ardavín C.
 Type I interferon (IFN) is crucial during infection through its antiviral properties and by coordinating the immunocompetent cells involved in antiviral or antibacterial immunity. Type I IFN (IFN-α and IFN-β) is produced after virus or bacteria recognition by cytosolic receptors or membrane-bound TLR receptors following the activation of the transcription factors IRF3 or IRF7. IFN-β production after fungal infection was recently reported, although the underlying mechanism remains controversial.
Type I interferon (IFN) is crucial during infection through its antiviral properties and by coordinating the immunocompetent cells involved in antiviral or antibacterial immunity. Type I IFN (IFN-α and IFN-β) is produced after virus or bacteria recognition by cytosolic receptors or membrane-bound TLR receptors following the activation of the transcription factors IRF3 or IRF7. IFN-β production after fungal infection was recently reported, although the underlying mechanism remains controversial.
Here we describe that IFN-β production by dendritic cells (DCs) induced by Candida albicans is largely dependent on Dectin-1- and Dectin-2-mediated signaling. Dectin-1-induced IFN-β production required the tyrosine kinase Syk and the transcription factor IRF5. Type I IFN receptor-deficient mice had a lower survival after C. albicans infection, paralleled by defective renal neutrophil infiltration. IFN-β production by renal infiltrating leukocytes was severely reduced in C. albicans-infected mice with Syk-deficient DCs. These data indicate that Dectin-induced IFN-β production by renal DCs is crucial for defense against C. albicans infection.
Cell Rep. 2013 Jul 2. pii: S2211-1247(13)00292-1
Gatchalian J, Fütterer A, Rothbart SB, Tong Q, Rincon-Arano H, Sánchez de Diego A, Groudine M, Strahl BD, Martínez-A C, van Wely KH, Kutateladze TG.

Binding of Dido3 PHD to histone H3K4me3 is disrupted by threonine phosphorylation that triggers Dido3 translocation from chromatin to the mitotic spindle. The crystal structure of Dido3 PHD in complex with H3K4me3 reveals an atypical aromatic-cage-like binding site that contains a histidine residue. Biochemical, structural, and mutational analyses of the binding mechanism identified the determinants of specificity and affinity and explained the inability of homologous PHF3 to bind H3K4me3. Together, our findings reveal a link between the transcriptional control in embryonic development and regulation of cell division.
PLoS One. 2013 Jun 27;8(6):e66894
García-Arriaza J, Arnáez P, Gómez CE, Sorzano CO, Esteban M.
Poxvirus vector Modified Vaccinia Virus Ankara (MVA) expressing HIV-1 Env, Gag, Pol and Nef antigens from clade B (termed MVA-B) is a promising HIV/AIDS vaccine candidate, as confirmed from results obtained in a prophylactic phase I clinical trial in humans. To improve the immunogenicity elicited by MVA-B, we have generated and characterized the innate immune sensing and the in vivo immunogenicity profile of a vector with a double deletion in two vaccinia virus (VACV) genes (C6L and K7R) coding for inhibitors of interferon (IFN) signaling pathways. The innate immune signals elicited by MVA-B deletion mutants (MVA-B ΔC6L and MVA-B ΔC6L/K7R) in human macrophages and monocyte-derived dendritic cells (moDCs) showed an up-regulation of the expression of IFN-β, IFN-α/β-inducible genes, TNF-α, and other cytokines and chemokines. A DNA prime/MVA boost immunization protocol in mice revealed that these MVA-B deletion mutants were able to improve the magnitude and quality of HIV-1-specific CD4+ and CD8+ T cell adaptive and memory immune responses, which were mostly mediated by CD8+ T cells of an effector phenotype, with MVA-B ΔC6L/K7R being the most immunogenic virus recombinant. CD4+ T cell responses were mainly directed against Env, while GPN-specific CD8+ T cell responses were induced preferentially by the MVA-B deletion mutants. Furthermore, antibody levels to Env in the memory phase were slightly enhanced by the MVA-B deletion mutants compared to the parental MVA-B.
These findings revealed that double deletion of VACV genes that act blocking intracellularly the IFN signaling pathway confers an immunological benefit, inducing innate immune responses and increases in the magnitude, quality and durability of the HIV-1-specific T cell immune responses. Our observations highlighted the immunomodulatory role of the VACV genes C6L and K7R, and that targeting common pathways, like IRF3/IFN-β signaling, could be a general strategy to improve the immunogenicity of poxvirus-based vaccine candidates.
PLoS One. 2013 Jun 14;8(6):e65526
Sanz-Sánchez L, Risco C.
Inside cells, viruses build specialized compartments for replication and morphogenesis. We observed that virus release associates with specific structures found on the surface of mammalian cells. Cultured adherent cells were infected with a bunyavirus and processed for oriented sectioning and transmission electron microscopy. Imaging of cell basal regions showed sophisticated multilamellar structures (MLS) and extracellular filament bundles with attached viruses.
Correlative light and electron microscopy confirmed that both MLS and filaments proliferated during the maximum egress of new viruses. MLS dimensions and structure were reminiscent of those reported for the nanostructures on gecko fingertips, which are responsible for the extraordinary attachment capacity of these lizards. As infected cells with MLS were more resistant to detachment than control cells, we propose an adhesive function for these structures, which would compensate for the loss of adherence during release of new virus progeny.
Produciéndose cerca de 10.000 alteraciones diarias en el genoma de cada célula, el trabajo que hacen proteínas reparadoras del ADN como las que estudia en su laboratorio del Centro Nacional de Biotecnología del CSIC (CNB) el grupo dirigido por el biofísico Fernando Moreno-Herrero es tremendamente importante.
 Junto a bioquímicos de la Universidad de Bristol, han publicado en la revista PNAS el mecanismo de exploración o escaneo que utiliza la proteína AddAB en el proceso de reparación de un corte de la doble hélice de ADN. Como nos explica Moreno-Herrero, reparar los daños en esencial ya que su acumulación puede acabar provocando "la muerte celular o incluso degenerar en una célula cancerígena".
Junto a bioquímicos de la Universidad de Bristol, han publicado en la revista PNAS el mecanismo de exploración o escaneo que utiliza la proteína AddAB en el proceso de reparación de un corte de la doble hélice de ADN. Como nos explica Moreno-Herrero, reparar los daños en esencial ya que su acumulación puede acabar provocando "la muerte celular o incluso degenerar en una célula cancerígena".
Con anterioridad, el grupo del CNB había demostrado que para que la proteína AddAB recorriera el cromosoma separando las dos hebras necesita estar unida a unas secuencias del genoma llamadas Chi. Ahora además, utilizando la técnica de pinzas magnéticas, han sido capaces de estudiar molécula a molécula cómo recorre el ADN, a qué velocidad y hasta las pausas que hacen. Sus resultados muestran que las características de la translocación a lo largo del ADN dependen del contenido en citosina y guanina y de la presencia de las secuencias Chi.
Desde el CNB, la investigadora postdoctoral Carolina Carrasco nos explica que en su estudio han podido ver "que cuando la proteína encuentra una verdadera secuencia Chi, realiza una pausa en esta posición después de la cual reinicia nuevamente su trayectoria". Y es que cuando AddAB se encuentra con secuencias parecidas a Chi, también se detiene en un intento fallido de reconocer estas pseudo-Chi, siendo la duración de las pausas en estos casos menores.
Basándose en la capacidad de dicha proteína de reconocer específicamente las secuencias Chi, los investigadores han propuesto un modelo mecánico para explicar el origen de ambas pausas. La forma de la pausa y el aparente cambio de velocidad después de la misma actúan cómo un filtro selectivo cuando se encuentra ante una secuencia Chi genuina.
- Carrasco C, Gilhooly NS, Dillingham MS, Moreno-Herrero F. On the mechanism of recombination hotspot scanning during double-stranded DNA break resection. PNAS June 24, 2013.
Carrasco C, Gilhooly NS, Dillingham MS, Moreno-Herrero F.
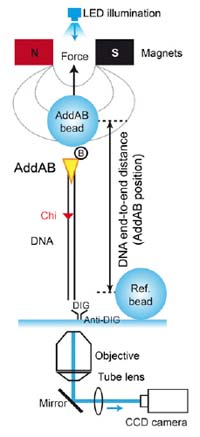 We report the use of the magnetic tweezers approach to monitor the dynamics of double-stranded DNA break resection at single-molecule resolution. In this work, we unveil the inner workings of the Bacillus subtilis AddAB complex by showing that the domain responsible for the recognition of the recombination hotspot sequence Crossover hotspot instigator (Chi) antagonizes the translocation activity, causing the complex to transiently stall at Chi and Chi-like sequences.
We report the use of the magnetic tweezers approach to monitor the dynamics of double-stranded DNA break resection at single-molecule resolution. In this work, we unveil the inner workings of the Bacillus subtilis AddAB complex by showing that the domain responsible for the recognition of the recombination hotspot sequence Crossover hotspot instigator (Chi) antagonizes the translocation activity, causing the complex to transiently stall at Chi and Chi-like sequences.
We propose a simple model for failed and successful Chi recognition in which the battle between the translocation and sequence-specific binding activities acts as a selectivity filter for bona fide Chi sequences.
 Vivimos rodeados de microbios. Microbios
buenos, microbios malos y, sobre todo, microbios ni buenos ni malos.
Vivimos rodeados de microbios. Microbios
buenos, microbios malos y, sobre todo, microbios ni buenos ni malos.
Las bacterias son un tipo de microbios tan pequeños que no los podemos ver a simple vista. Ni siquiera usando una lupa. Para poder verlas necesitamos un microscopio. Por eso, hasta que no se inventó el microscopio nadie sabía que existían.
Ahora sabemos que hay un montón de tipos distintos y conocemos muchas de las que viven cerca de nosotros: en el agua, en la tierra o dentro de nosotros mismos. Sí, incluso dentro de nuestras barrigas. Son minúsculas, pero todas juntas pesan casi tanto como un litro de leche.
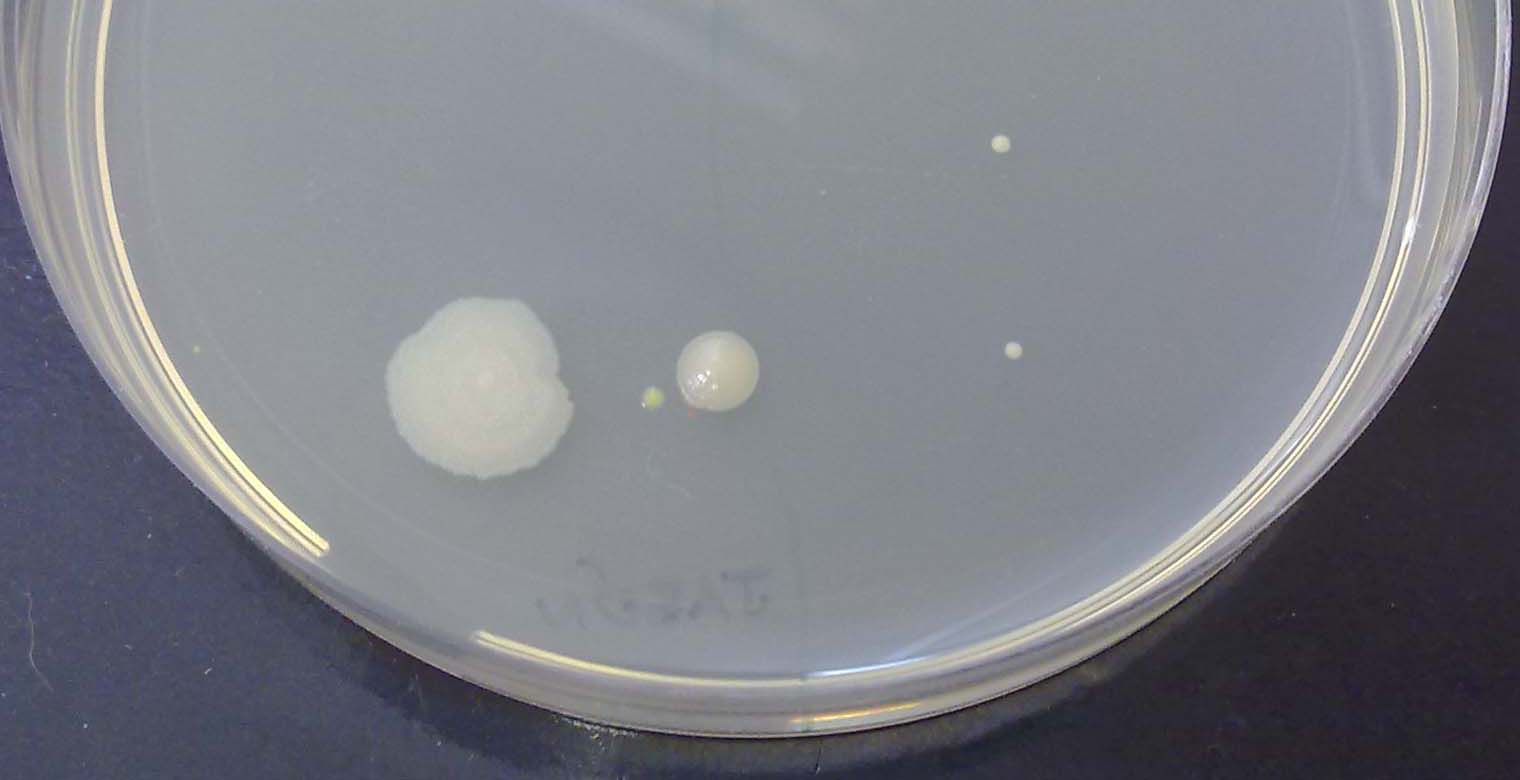
Aunque no las podamos ver una a una, los científicos saben criarlas y tener tal cantidad de ellas que podemos ver los grupos que forman. A estos grupos, los científicos los llaman colonias.
Un científico llamado Julius Richard Petri inventó un sistema muy sencillo para criar las bacterias: una placa redonda en la que hay una gelatina que tiene los alimentos que las bacterias necesitan. ¿Cuántos grupos de bacterias (colonias) ves que han crecido en esta placa?.
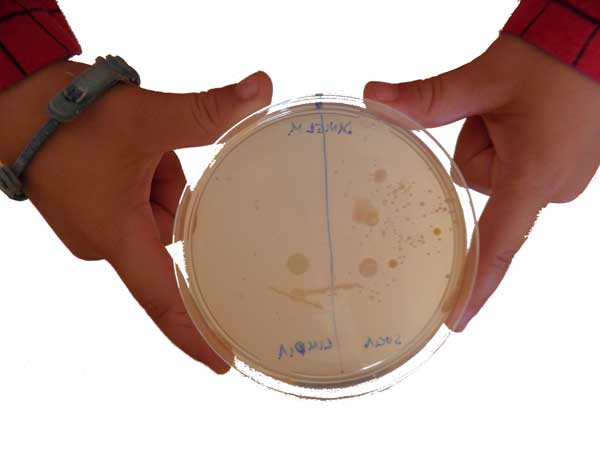
En nuestras manos podemos encontrar un montón de tipos distintos de bacterias. Para verlas, tan solo tenemos que tocar la gelatina de una de las placas de Petri y dejar que crezcan las colonias durante unos días. ¡Tócala con cuidado, no vaya a ser que se rompa!
Si lo haces con las manos limpias, ¿habrá más o menos bacterias? ¿Te apetece hacer el experimento? Toca una mitad de la placa con las manos sucias. Luego lávate las manos y toca la otra mitad. Según pasen los días, observa cómo crecen las colonias.
Como un buen científico, ve anotando durante los días siguientes cuantas colonias salen con las manos sucias y cuantas con ellas limpias.
El último día compara tus resultados con los del resto de la clase y pensar entre todos qué les pasa a las bacterias cuando te lavas las manos.
Una vez que hayan terminado todos los grupos, hay en poner en común los resultados obtenidos y sumar los números de todos y cada una de las parejas.
Esta práctica está basada en una visita que nos hicieron a principios de este año los alumnos del Colegio Rafael Alberti de Coslada. Si te interesa, esta sencilla práctica dirigida a alumnos educación primaria se puede descargar en pdf (2,37 MB).
COOKIES POLICY
A cookie is a text file that is stored on your computer or mobile device via a web server and only that server will be able to retrieve or read the contents of the cookie and allow the Web site remember browser preferences and navigate efficiently. Cookies make the interaction between the user and the website faster and easier.
General information
This Website uses cookies. Cookies are small text files generated by the web pages you visit, which contain the session data that can be useful later in the website. In this way this Web remembers information about your visit, which can facilitate your next visit and make the website more useful.
How do cookies?
Cookies can only store text, usually always anonymous and encrypted. No personal information is ever stored in a cookie, or can be associated with identified or identifiable person.
The data allow this website to keep your information between the pages, and also to discuss how to interact with the website. Cookies are safe because they can only store information that is put there by the browser, which is information the user entered in the browser or included in the page request. You can not run the code and can not be used to access your computer. If a website encrypts cookie data, only the website can read the information.
What types of cookies used?
The cookies used by this website can be distinguished by the following criteria:
1. Types of cookies as the entity that manages:
Depending on who the entity operating the computer or domain where cookies are sent and treat the data obtained, we can distinguish:
- Own cookies: are those that are sent to the user's terminal equipment from a computer or domain managed by the editor itself and from which provides the service requested by the user.
- Third party cookies: these are those that are sent to the user's terminal equipment from a machine or domain that is not managed by the publisher, but by another entity data is obtained through cookies.
In the event that the cookies are installed from a computer or domain managed by the editor itself but the information collected by these is managed by a third party can not be considered as party cookies.
2. Types of cookies as the length of time that remain active:
Depending on the length of time that remain active in the terminal equipment can be distinguished:
- Session cookies: cookies are a type designed to collect and store data while the user accesses a web page. Are usually used to store information that only worth preserving for the service requested by the user at any one time (eg a list of products purchased).
- Persistent cookies: cookies are a type of data which are stored in the terminal and can be accessed and treated for a period defined by the head of the cookie, and can range from a few minutes to several years.
3. Cookies types according to their purpose:
Depending on the purpose for which the data are processed through cookies, we can distinguish between:
- Technical cookies: these are those that allow the user to navigate through a web page or application platform and the use of different options or services it exist as, for example, control traffic and data communication, identify the session, access to restricted access parts, remember the elements of an order, make the buying process an order, make an application for registration or participation in an event, use security features while browsing store content for dissemination videos or sound or share content via social networks.
- Customization cookies: these are those that allow the user to access the service with some general characteristics based on a predefined set of criteria in the user terminal would eg language, the type of browser through which you access the service, the locale from which you access the service, etc.
- Analysis cookies: they are those that allow the responsible for them, monitoring and analyzing the behavior of users of the web sites that are linked. The information gathered through such cookies are used in measuring the activity of web sites, application or platform and for the profiling of user navigation of such sites, applications and platforms, in order to make improvements function data analysis how users use the service.
Management tool cookies
This Website uses Google Analytics.
Google Analytics is a free tool from Google that primarily allows website owners know how users interact with your website. Also, enable cookies in the domain of the site in which you are and uses a set of cookies called "__utma" and "__utmz" to collect information anonymously and reporting of website trends without identifying individual users..
For statistics of use of this website use cookies in order to know the level of recurrence of our visitors and more interesting content. This way we can concentrate our efforts on improving the most visited areas and make the user more easily find what they are looking for. On this site you can use the information from your visit for statistical evaluations and calculations anonymous data and to ensure the continuity of service or to make improvements to their websites. For more details, see the link below privacy policy [http://www.google.com/intl/en/policies/privacy/]
How to manage cookies on your computer: disabling and deleting cookies
All Internet browsers allow you to limit the behavior of a cookie or disable cookies within settings or browser settings. The steps for doing so are different for each browser, you can find instructions in the help menu of your browser.
If you decline the use of cookies, since it is possible thanks to the preferences menu of your browser or settings, reject, this website will continue to function properly without the use of the same.
Can you allow, block or delete cookies installed on your computer by setting your browser options installed on your computer:
- For more information about Internet Explorer click here.
- For more information on Chrome click here.
- For more information about Safari click here.
- For more information about Firefox click here.
Through your browser, you can also view the cookies that are on your computer, and delete them as you see fit. Cookies are text files, you can open and read the contents. The data within them is almost always encrypted with a numeric key corresponding to an Internet session so often has no meaning beyond the website who wrote it.
Informed consent
The use of this website on the other hand, implies that you paid your specific consent to the use of cookies, on the terms and conditions provided in this Cookies Policy, without prejudice to the measures of deactivation and removal of cookies that you can take, and mentioned in the previous section.





