J Am Chem Soc. 2013 Aug 21;135(33):12366-76
Blanco B, Prado V, Lence E, Otero JM, Garcia-Doval C, van Raaij MJ, Llamas-Saiz AL, Lamb H, Hawkins AR, González-Bello C.
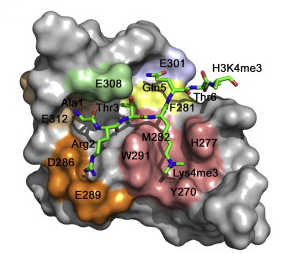
 Shikimate kinase (SK) is an essential enzyme in several pathogenic bacteria and does not have any counterpart in human cells, thus making it an attractive target for the development of new antibiotics. The key interactions of the substrate and product binding and the enzyme movements that are essential for catalytic turnover of the Mycobacterium tuberculosis shikimate kinase enzyme (Mt-SK) have been investigated by structural and computational studies. Based on these studies several substrate analogs were designed and assayed.
Shikimate kinase (SK) is an essential enzyme in several pathogenic bacteria and does not have any counterpart in human cells, thus making it an attractive target for the development of new antibiotics. The key interactions of the substrate and product binding and the enzyme movements that are essential for catalytic turnover of the Mycobacterium tuberculosis shikimate kinase enzyme (Mt-SK) have been investigated by structural and computational studies. Based on these studies several substrate analogs were designed and assayed.
The crystal structure of Mt-SK in complex with ADP and one of the most potent inhibitors has been solved at 2.15 Å. These studies reveal that the fixation of the diaxial conformation of the C4 and C5 hydroxyl groups recognized by the enzyme or the replacement of the C3 hydroxyl group in the natural substrate by an amino group is a promising strategy for inhibition because it causes a dramatic reduction of the flexibility of the LID and shikimic acid binding domains. Molecular dynamics simulation studies showed that the product is expelled from the active site by three arginines (Arg117, Arg136, and Arg58). This finding represents a previously unknown key role of these conserved residues. These studies highlight the key role of the shikimic acid binding domain in the catalysis and provide guidance for future inhibitor designs.
El próximo 4 de octubre de 2013 comienza el XVIII Ciclo de Seminarios del CNB con una conferencia impartida por Matteo Iannacone.
Con la colaboración de Sigma-Aldrich, AppliChem, VWR y Panreac, a lo largo del curso 2013-2014 contaremos con la presencia de 13 magníficos científicos que explicarán sus últimas investigaciones en el campo de la Ciencias de la vida.
A execpción de la ponencia de Philip Cohen que será un jueves, el resto tendrá lugar los viernes a las 12:00 en el Salón de Actos del Centro Nacional de Biotecnología del CSIC.
PLoS One. 2013 Aug 7;8(8):e70364
Banon-Rodriguez I, Saez de Guinoa J, Bernardini A, Ragazzini C, Fernandez E, Carrasco YR, Jones GE, Wandosell F, Anton IM.
The spatial distribution of signals downstream from receptor tyrosine kinases (RTKs) or G-protein coupled receptors (GPCR) regulates fundamental cellular processes that control cell migration and growth. Both pathways rely significantly on actin cytoskeleton reorganization mediated by nucleation-promoting factors such as the WASP-(Wiskott-Aldrich Syndrome Protein) family. WIP (WASP Interacting Protein) is essential for the formation of a class of polarised actin microdomain, namely dorsal ruffles, downstream of the RTK for PDGF (platelet-derived growth factor) but the underlying mechanism is poorly understood.
Using lentivirally-reconstituted WIP-deficient murine fibroblasts we define the requirement for WIP interaction with N-WASP (neural WASP) and Nck for efficient dorsal ruffle formation and of WIP-Nck binding for fibroblast chemotaxis towards PDGF-AA. The formation of both circular dorsal ruffles in PDGF-AA-stimulated primary fibroblasts and lamellipodia in CXCL13-treated B lymphocytes are also compromised by WIP-deficiency. We provide data to show that a WIP-Nck signalling complex interacts with RTK to promote polarised actin remodelling in fibroblasts and provide the first evidence for WIP involvement in the control of migratory persistence in both mesenchymal (fibroblast) and amoeboid (B lymphocytes) motility.
Las células "madre" embrionarias reciben este nombre por su capacidad de generar cualquier tipo celular de las tres capas del embrión a partir de las cuales se originarán los tejidos y órganos en el individuo adulto: el endodermo, el mesodermo o el ectodermo (diferenciación). Además, y al contrario que las células adultas especializadas, las células madre tienen la asombrosa capacidad de poder multiplicarse indefinidamente en el laboratorio (división), lo que las hace tremendamente útiles en el campo de la medicina regenerativa.
 En la actualidad, los científicos tratan de comprender los mecanismos moleculares que regulan en las células madre el equilibrio entre ambos procesos, la división y la diferenciación. Últimamente se han identificado genes que permiten reprogramar células especializadas y convertirlas en células madre pero aun hay muchos genes específicos de estas células cuya función es desconocida.
En la actualidad, los científicos tratan de comprender los mecanismos moleculares que regulan en las células madre el equilibrio entre ambos procesos, la división y la diferenciación. Últimamente se han identificado genes que permiten reprogramar células especializadas y convertirlas en células madre pero aun hay muchos genes específicos de estas células cuya función es desconocida.
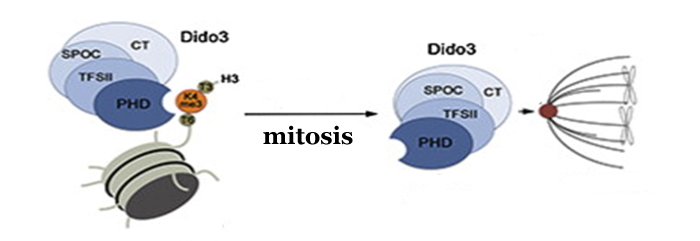
Un ejemplo es el gen Dido al que dedica sus esfuerzos el grupo del Centro Nacional de Biotecnología del CSIC (CNB) dirigido por Carlos Martínez-A. Este gen aparece evolutivamente en vertebrados y se manifiesta en tres proteínas distintas: Dido1, Dido2 y Dido3. Esta última se expresa en células madre y se modula su expresión en las células especializadas. Se une a estructuras esenciales para la división celular como son el centrosoma y el complejo sinaptonémico. Alteraciones de esta proteína causan síndromes preleucémicos y tumorales y bloquean la capacidad de diferenciarse de las células madre en cualquier tipo celular específico (diferenciación) manteniendo su capacidad de división (proliferación).
En colaboración con investigadores de la Universidad de Colorado, los científicos del CNB acaban de publicar un trabajo en el que identifican al dominio PH de Dido3, y determinan su estructura cristalina a 1.8 A de resolución, como el responsable de la unión al cromosoma. Esta unión causaría modificaciones en la expresión de diferentes genes que le dan a las células madre su capacidad de dividirse y diferenciarse en células adultas.
Además de identificar la zona de la proteína que se une a las histonas que organizan los cromosomas, los investigadores han profundizado en cómo se regula su función según el momento del ciclo celular en que se encuentre la célula. Sus datos indican que cuando las células madre se diferencian, Dido3 es desplazada del cromosoma por el aumento de Dido1. Esto conlleva una disminución en la expresión de genes que dotan a las células madre de su pluripotencia.
En este mismo aspecto, durante la mitosis han observado que Dido3 pasa de estar unido a la cromatina a desplazarse hasta el huso mitótico que dispone a los cromosomas en el ecuador de la célula como paso previo a separarlos hacia los polos cuando se va a producir la división de una célula en dos. De este modo, queda descrita por primera vez a través de Dido relación entre la expresión de genes durante el desarrollo embrionario y la regulación del ciclo celular
- Gatchalian J, Fütterer A, Rothbart SB, Tong Q, Rincon-Arano H, Sánchez de Diego A, Groudine M, Strahl BD, Martínez-A C, van Wely KH, Kutateladze TG. Dido3 PHD modulates cell differentiation and division. Cell Rep 2013 Jul 2. pii: S2211-1247(13)00292-1.
Environ Microbiol. 2013 Aug 1. doi: 10.1111/1462-2920.12225
Pazos M, Natale P, Margolin W, Vicente M.
 We used bimolecular fluorescence complementation (BiFC) assays to detect protein-protein interactions of all possible pairs of the essential Escherichia coli proto-ring components, FtsZ, FtsA and ZipA, as well as the non-essential FtsZ-associated proteins ZapA and ZapB. We found an unexpected interaction between ZipA and ZapB at potential cell division sites, and when co-overproduced, they induced long narrow constrictions at division sites that were dependent on FtsZ.
We used bimolecular fluorescence complementation (BiFC) assays to detect protein-protein interactions of all possible pairs of the essential Escherichia coli proto-ring components, FtsZ, FtsA and ZipA, as well as the non-essential FtsZ-associated proteins ZapA and ZapB. We found an unexpected interaction between ZipA and ZapB at potential cell division sites, and when co-overproduced, they induced long narrow constrictions at division sites that were dependent on FtsZ.
These assays also uncovered an interaction between ZipA and ZapA that was mediated by FtsZ. BiFC with ZapA and ZapB showed that in addition to their expected interaction at midcell, they also interact at the cell poles. BiFC detected interaction between FtsZ and ZapB at midcell and close to the poles. Results from the remaining pairwise combinations confirmed known interactions between FtsZ and ZipA, and ZapB with itself.
J Biol Chem. 2013 Jun 7;288(23):16998-7007
Daudén MI, Martín-Benito J, Sánchez-Ferrero JC, Pulido-Cid M, Valpuesta JM, Carrascosa JL.
 During bacteriophage morphogenesis DNA is translocated into a preformed prohead by the complex formed by the portal protein, or connector, plus the terminase, which are located at an especial prohead vertex. The terminase is a powerful motor that converts ATP hydrolysis into mechanical movement of the DNA. Here, we have determined the structure of the T7 large terminase by electron microscopy.
During bacteriophage morphogenesis DNA is translocated into a preformed prohead by the complex formed by the portal protein, or connector, plus the terminase, which are located at an especial prohead vertex. The terminase is a powerful motor that converts ATP hydrolysis into mechanical movement of the DNA. Here, we have determined the structure of the T7 large terminase by electron microscopy.
The five terminase subunits assemble in a toroid that encloses a channel wide enough to accommodate dsDNA. The structure of the complete connector-terminase complex is also reported, revealing the coupling between the terminase and the connector forming a continuous channel. The structure of the terminase assembled into the complex showed a different conformation when compared with the isolated terminase pentamer. To understand in molecular terms the terminase morphological change, we generated the terminase atomic model based on the crystallographic structure of its phage T4 counterpart. The docking of the threaded model in both terminase conformations showed that the transition between the two states can be achieved by rigid body subunit rotation in the pentameric assembly. The existence of two terminase conformations and its possible relation to the sequential DNA translocation may shed light into the molecular bases of the packaging mechanism of bacteriophage T7.
J Struct Biol. 2013 Aug 6. pii: S1047-8477(13)00195-0
Vargas J, Abrishami V, Marabini R, de la Rosa-Trevín JM, Zaldivar A, Carazo JM, Sorzano CO.
 Three-dimensional reconstruction of biological specimens using electron microscopy by single particle methodologies requires the identification and extraction of the imaged particles from the acquired micrographs. Automatic and semiautomatic particle selection approaches can localize these particles, minimizing the user interaction, but at the cost of selecting a non-negligible number of incorrect particles, which can corrupt the final three-dimensional reconstruction.
Three-dimensional reconstruction of biological specimens using electron microscopy by single particle methodologies requires the identification and extraction of the imaged particles from the acquired micrographs. Automatic and semiautomatic particle selection approaches can localize these particles, minimizing the user interaction, but at the cost of selecting a non-negligible number of incorrect particles, which can corrupt the final three-dimensional reconstruction.
In this work, we present a novel particle quality assessment and sorting method that can separate most erroneously picked particles from correct ones. The proposed method is based on multivariate statistical analysis of a particle set that has been picked previously using any automatic or manual approach. The new method uses different sets of particle descriptors, which are morphology-based, histogram-based and signal to noise analysis based. We have tested our proposed algorithm with experimental data obtaining very satisfactory results. The algorithm is freely available as a part of the Xmipp 3.0 package.
La presencia de arsénico en la naturaleza debido a las erupciones volcánicas ha sido una de las amenazas más importantes para el desarrollo de la vida en la tierra. Hoy en día el arsénico presente en los suelos, puede ser solubilizado en las aguas subterráneas exponiendo a los organismos a uno de los carcinógenos más potentes que se conocen. De hecho la exposición a arsénico es responsable del envenenamiento masivo más importante que ha sufrido la humanidad en toda su historia.
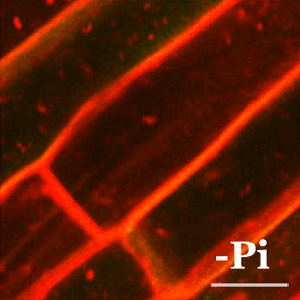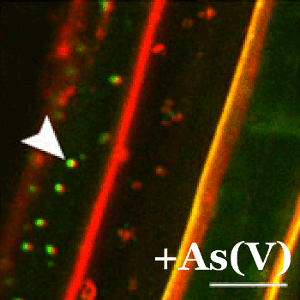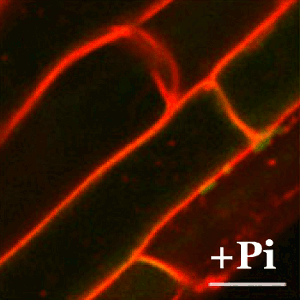 En plantas, la tolerancia al arsénico es esencial para su supervivencia ya que la forma química más abundante en la tierra (el arseniato) guarda una gran similitud con el fosfato y las plantas lo incorporan fácilmente a sus células a través de los transportadores de fosfato. En el CNB, el grupo dirigido por Antonio Leyva estudia las bases moleculares de los mecanismos de percepción de arsénico en plantas con el fin de identificar y desarrollar plantas que eviten la exposición de este carcinógeno a los seres vivos.
En plantas, la tolerancia al arsénico es esencial para su supervivencia ya que la forma química más abundante en la tierra (el arseniato) guarda una gran similitud con el fosfato y las plantas lo incorporan fácilmente a sus células a través de los transportadores de fosfato. En el CNB, el grupo dirigido por Antonio Leyva estudia las bases moleculares de los mecanismos de percepción de arsénico en plantas con el fin de identificar y desarrollar plantas que eviten la exposición de este carcinógeno a los seres vivos.
En este trabajo, merecedor de un comentario especial por parte de los editores de la revista Plant Cell, el grupo de Leyva ha descubierto que cuando las plantas detectan el arseniato impiden de forma inmediata su captación mediante la represión y deslocalización del transportador de fosfato. Este sistema está controlado mediante la acción de una proteína represora conocida como WRKY6.
En colaboración con otros dos grupos del CNB, han podido además descubrir que el arseniato provoca la activación masiva de transposones, unos elementos del ADN móviles capaces de “saltar” de un sitio a otro del genoma en respuesta a estrés y por tanto actuar como agentes mutagénicos. Una activación que también está controlada por el represor WRKY6.
Estos resultados indican que las plantas poseen un mecanismo de percepción del arseniato que controla la activación de transposones y la incorporación del arseniato. Todo ello proporciona un sistema integrado de tolerancia y de estabilidad genómica.
- Castrillo G, Sánchez-Bermejo E, de Lorenzo L, Crevillén P, Fraile-Escanciano A, Tc M, Mouriz A, Catarecha P, Sobrino-Plata J, Olsson S, Leo Del Puerto Y, Mateos I, Rojo E, Hernández LE, Jarillo JA, Piñeiro M, Paz-Ares J, Leyva A. WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis. Plant Cell 2013 Aug 6 tpc.113.114009.
Curr Proteomics. 2013; 10(2):83-97
Casado-Vela J, González-González M, Matarraz S, Martínez-Esteso MJ, Vilella M, Sayagues JM, FuentesM, Lacal JC.
 Protein arrays represent a step forward in proteomic technologies, as they enable to address biological and functional questions by large-scale approaches (i.e., up to several thousand proteins may be monitored in a single experiment). Here we review recent achievements of protein arrays and their classification. We also summarize current and future applications of this technology to address the study of the human proteome including protein identification, biomarker discovery, screening of protein-protein/molecule interactions, detection of protein post-translational modifications and serum profiling.
Protein arrays represent a step forward in proteomic technologies, as they enable to address biological and functional questions by large-scale approaches (i.e., up to several thousand proteins may be monitored in a single experiment). Here we review recent achievements of protein arrays and their classification. We also summarize current and future applications of this technology to address the study of the human proteome including protein identification, biomarker discovery, screening of protein-protein/molecule interactions, detection of protein post-translational modifications and serum profiling.
Advantages and limitations of protein arrays compared to other proteomic technologies are also discussed here. The use, advances and recent applications of fluorescent microspheres as a promising alternative to planar arrays are also summarized in this report. Finally, we propose future perspectives of protein arrays from a comprehensive standpoint that includes technological, biological, economical and practical aspects.
Nikel PI, de Lorenzo V.
 Pseudomonas putida KT2440, a microbial cell factory of reference for industrial whole-cell biocatalysis, is unable to support biochemical reactions that occur under anoxic conditions, limiting its utility for a large number of relevant biotransformations. Unlike (facultative) anaerobes, P. putida resorts to NADH oxidation via an oxic respiratory chain and completely lacks a true fermentation metabolism. Therefore, it cannot achieve the correct balances of energy and redox couples (i.e., ATP/ADP and NADH/NAD+) that are required to sustain an O2-free lifestyle.
Pseudomonas putida KT2440, a microbial cell factory of reference for industrial whole-cell biocatalysis, is unable to support biochemical reactions that occur under anoxic conditions, limiting its utility for a large number of relevant biotransformations. Unlike (facultative) anaerobes, P. putida resorts to NADH oxidation via an oxic respiratory chain and completely lacks a true fermentation metabolism. Therefore, it cannot achieve the correct balances of energy and redox couples (i.e., ATP/ADP and NADH/NAD+) that are required to sustain an O2-free lifestyle.
To overcome this state of affairs, the acetate kinase (ackA) gene of the facultative anaerobe Escherichia coli and the pyruvate decarboxylase (pdc) and alcohol dehydrogenase II (adhB) genes of the aerotolerant Zymomonas mobilis were knocked-in to a wild-type P. putida strain. Biochemical and genetic assays showed that conditional expression of the entire enzyme set allowed the engineered bacteria to adopt an anoxic regime that maintained considerable metabolic activity. The resulting strain was exploited as a host for the heterologous expression of a 1,3-dichloroprop-1-ene degradation pathway recruited from Pseudomonas pavonaceae 170, enabling the recombinants to degrade this recalcitrant chlorinated compound anoxically. These results underscore the value of P. putida as a versatile agent for biotransformations able to function at progressively lower redox statuses.
COOKIES POLICY
A cookie is a text file that is stored on your computer or mobile device via a web server and only that server will be able to retrieve or read the contents of the cookie and allow the Web site remember browser preferences and navigate efficiently. Cookies make the interaction between the user and the website faster and easier.
General information
This Website uses cookies. Cookies are small text files generated by the web pages you visit, which contain the session data that can be useful later in the website. In this way this Web remembers information about your visit, which can facilitate your next visit and make the website more useful.
How do cookies?
Cookies can only store text, usually always anonymous and encrypted. No personal information is ever stored in a cookie, or can be associated with identified or identifiable person.
The data allow this website to keep your information between the pages, and also to discuss how to interact with the website. Cookies are safe because they can only store information that is put there by the browser, which is information the user entered in the browser or included in the page request. You can not run the code and can not be used to access your computer. If a website encrypts cookie data, only the website can read the information.
What types of cookies used?
The cookies used by this website can be distinguished by the following criteria:
1. Types of cookies as the entity that manages:
Depending on who the entity operating the computer or domain where cookies are sent and treat the data obtained, we can distinguish:
- Own cookies: are those that are sent to the user's terminal equipment from a computer or domain managed by the editor itself and from which provides the service requested by the user.
- Third party cookies: these are those that are sent to the user's terminal equipment from a machine or domain that is not managed by the publisher, but by another entity data is obtained through cookies.
In the event that the cookies are installed from a computer or domain managed by the editor itself but the information collected by these is managed by a third party can not be considered as party cookies.
2. Types of cookies as the length of time that remain active:
Depending on the length of time that remain active in the terminal equipment can be distinguished:
- Session cookies: cookies are a type designed to collect and store data while the user accesses a web page. Are usually used to store information that only worth preserving for the service requested by the user at any one time (eg a list of products purchased).
- Persistent cookies: cookies are a type of data which are stored in the terminal and can be accessed and treated for a period defined by the head of the cookie, and can range from a few minutes to several years.
3. Cookies types according to their purpose:
Depending on the purpose for which the data are processed through cookies, we can distinguish between:
- Technical cookies: these are those that allow the user to navigate through a web page or application platform and the use of different options or services it exist as, for example, control traffic and data communication, identify the session, access to restricted access parts, remember the elements of an order, make the buying process an order, make an application for registration or participation in an event, use security features while browsing store content for dissemination videos or sound or share content via social networks.
- Customization cookies: these are those that allow the user to access the service with some general characteristics based on a predefined set of criteria in the user terminal would eg language, the type of browser through which you access the service, the locale from which you access the service, etc.
- Analysis cookies: they are those that allow the responsible for them, monitoring and analyzing the behavior of users of the web sites that are linked. The information gathered through such cookies are used in measuring the activity of web sites, application or platform and for the profiling of user navigation of such sites, applications and platforms, in order to make improvements function data analysis how users use the service.
Management tool cookies
This Website uses Google Analytics.
Google Analytics is a free tool from Google that primarily allows website owners know how users interact with your website. Also, enable cookies in the domain of the site in which you are and uses a set of cookies called "__utma" and "__utmz" to collect information anonymously and reporting of website trends without identifying individual users..
For statistics of use of this website use cookies in order to know the level of recurrence of our visitors and more interesting content. This way we can concentrate our efforts on improving the most visited areas and make the user more easily find what they are looking for. On this site you can use the information from your visit for statistical evaluations and calculations anonymous data and to ensure the continuity of service or to make improvements to their websites. For more details, see the link below privacy policy [http://www.google.com/intl/en/policies/privacy/]
How to manage cookies on your computer: disabling and deleting cookies
All Internet browsers allow you to limit the behavior of a cookie or disable cookies within settings or browser settings. The steps for doing so are different for each browser, you can find instructions in the help menu of your browser.
If you decline the use of cookies, since it is possible thanks to the preferences menu of your browser or settings, reject, this website will continue to function properly without the use of the same.
Can you allow, block or delete cookies installed on your computer by setting your browser options installed on your computer:
- For more information about Internet Explorer click here.
- For more information on Chrome click here.
- For more information about Safari click here.
- For more information about Firefox click here.
Through your browser, you can also view the cookies that are on your computer, and delete them as you see fit. Cookies are text files, you can open and read the contents. The data within them is almost always encrypted with a numeric key corresponding to an Internet session so often has no meaning beyond the website who wrote it.
Informed consent
The use of this website on the other hand, implies that you paid your specific consent to the use of cookies, on the terms and conditions provided in this Cookies Policy, without prejudice to the measures of deactivation and removal of cookies that you can take, and mentioned in the previous section.






