Carrasco C, Gilhooly NS, Dillingham MS, Moreno-Herrero F.
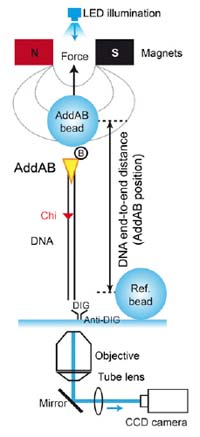 We report the use of the magnetic tweezers approach to monitor the dynamics of double-stranded DNA break resection at single-molecule resolution. In this work, we unveil the inner workings of the Bacillus subtilis AddAB complex by showing that the domain responsible for the recognition of the recombination hotspot sequence Crossover hotspot instigator (Chi) antagonizes the translocation activity, causing the complex to transiently stall at Chi and Chi-like sequences.
We report the use of the magnetic tweezers approach to monitor the dynamics of double-stranded DNA break resection at single-molecule resolution. In this work, we unveil the inner workings of the Bacillus subtilis AddAB complex by showing that the domain responsible for the recognition of the recombination hotspot sequence Crossover hotspot instigator (Chi) antagonizes the translocation activity, causing the complex to transiently stall at Chi and Chi-like sequences.
We propose a simple model for failed and successful Chi recognition in which the battle between the translocation and sequence-specific binding activities acts as a selectivity filter for bona fide Chi sequences.
 Vivimos rodeados de microbios. Microbios
buenos, microbios malos y, sobre todo, microbios ni buenos ni malos.
Vivimos rodeados de microbios. Microbios
buenos, microbios malos y, sobre todo, microbios ni buenos ni malos.
Las bacterias son un tipo de microbios tan pequeños que no los podemos ver a simple vista. Ni siquiera usando una lupa. Para poder verlas necesitamos un microscopio. Por eso, hasta que no se inventó el microscopio nadie sabía que existían.
Ahora sabemos que hay un montón de tipos distintos y conocemos muchas de las que viven cerca de nosotros: en el agua, en la tierra o dentro de nosotros mismos. Sí, incluso dentro de nuestras barrigas. Son minúsculas, pero todas juntas pesan casi tanto como un litro de leche.
Aunque no las podamos ver una a una, los científicos saben criarlas y tener tal cantidad de ellas que podemos ver los grupos que forman. A estos grupos, los científicos los llaman colonias.
Un científico llamado Julius Richard Petri inventó un sistema muy sencillo para criar las bacterias: una placa redonda en la que hay una gelatina que tiene los alimentos que las bacterias necesitan. ¿Cuántos grupos de bacterias (colonias) ves que han crecido en esta placa?.
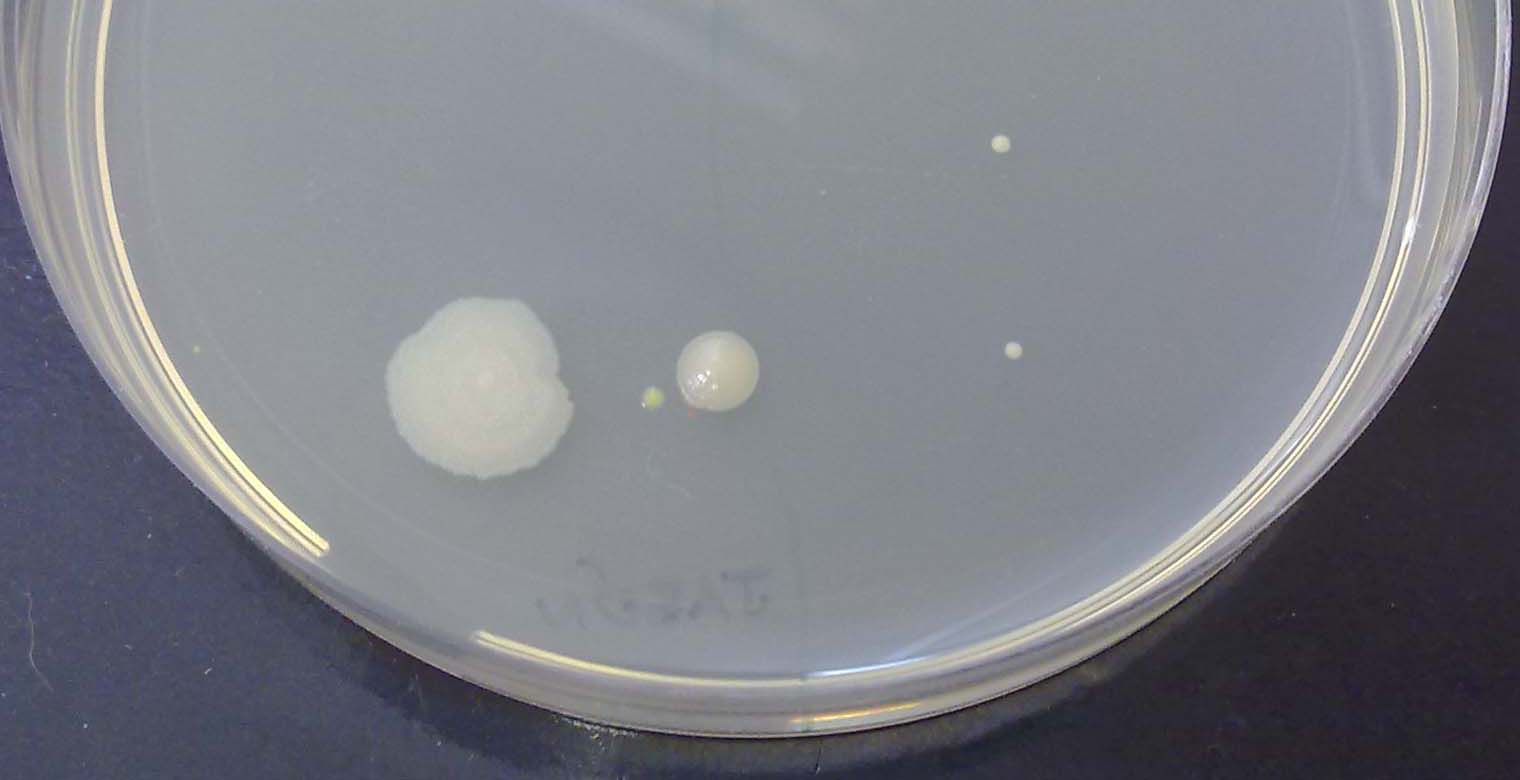
En nuestras manos podemos encontrar un montón de tipos distintos de bacterias. Para verlas, tan solo tenemos que tocar la gelatina de una de las placas de Petri y dejar que crezcan las colonias durante unos días. ¡Tócala con cuidado, no vaya a ser que se rompa!
Si lo haces con las manos limpias, ¿habrá más o menos bacterias? ¿Te apetece hacer el experimento? Toca una mitad de la placa con las manos sucias. Luego lávate las manos y toca la otra mitad. Según pasen los días, observa cómo crecen las colonias.
Como un buen científico, ve anotando durante los días siguientes cuantas colonias salen con las manos sucias y cuantas con ellas limpias.
El último día compara tus resultados con los del resto de la clase y pensar entre todos qué les pasa a las bacterias cuando te lavas las manos.
Una vez que hayan terminado todos los grupos, hay en poner en común los resultados obtenidos y sumar los números de todos y cada una de las parejas.
Esta práctica está basada en una visita que nos hicieron a principios de este año los alumnos del Colegio Rafael Alberti de Coslada. Si te interesa, esta sencilla práctica dirigida a alumnos educación primaria se puede descargar en pdf (2,37 MB).
Mol Cell. 2013 Jun 4. pii: S1097-2765(13)00365-1
Trinidad AG, Muller PA, Cuellar J, Klejnot M, Nobis M, Valpuesta JM, Vousden KH.

We show that folding of wild-type p53 is promoted by an interaction with the chaperonin CCT. Depletion of this chaperone in cells results in the accumulation of misfolded p53, leading to a reduction in p53-dependent gene expression. Intriguingly, p53 proteins mutated to prevent the interaction with CCT show conformational instability and acquire an ability to promote invasion and random motility that is similar to the activity of tumor-derived p53 mutants. Our data therefore suggest that both growth suppression and cell invasion may be differentially regulated functions of wild-type p53.
Ortega-Esteban A, Pérez-Berná AJ, Menéndez-Conejero R, Flint SJ, San Martín C, de Pablo PJ.
 The standard pathway for virus infection of eukaryotic cells requires disassembly of the viral shell to facilitate release of the viral genome into the host cell. Here we use mechanical fatigue, well below rupture strength, to induce stepwise disruption of individual human adenovirus particles under physiological conditions, and simultaneously monitor disassembly in real time.
The standard pathway for virus infection of eukaryotic cells requires disassembly of the viral shell to facilitate release of the viral genome into the host cell. Here we use mechanical fatigue, well below rupture strength, to induce stepwise disruption of individual human adenovirus particles under physiological conditions, and simultaneously monitor disassembly in real time.
Our data show the sequence of dismantling events in individual mature (infectious) and immature (noninfectious) virions, starting with consecutive release of vertex structures followed by capsid cracking and core exposure. Further, our experiments demonstrate that vertex resilience depends inextricably on maturation, and establish the relevance of penton vacancies as seeding loci for virus shell disruption. The mechanical fatigue disruption route recapitulates the adenovirus disassembly pathway in vivo, as well as the stability differences between mature and immature virions.
Por sexto año consecutivo, la Fundación "la Caixa" convoca su Programa Internacional de doctorado en Biomedicina en el Centro Nacional de Biotecnología del CSIC.
 Por su presupuesto y volumen de actividad, la Fundación "la Caixa" figura entre las diez fundaciones más importantes del mundo. Desde 1982, mantiene diversos programas para cursar estudios de postgrado tanto en España como en el extranjero.
Por su presupuesto y volumen de actividad, la Fundación "la Caixa" figura entre las diez fundaciones más importantes del mundo. Desde 1982, mantiene diversos programas para cursar estudios de postgrado tanto en España como en el extranjero.
En este sexto año consecutivo, la Fundación "la Caixa" convoca dos contratos predoctorales de personal investigador en formación para desarrollar un doctorado en Biomedicina en el Centro Nacional de Biotecnología del CSIC, el mayor centro español y uno de los centros líderes en el mundo en el área de Biotecnología, Biología Molecular y Biomedicina.
Las bases de la convocatoria de este año, publicadas en el Boletín Oficial del Estado del 24 de junio de 2013, pueden descargarse como documento pdf (4,61 MB).
Los estudiantes deberán aportar la documentación acreditativa de estar admitido en un programa de doctorado expedido por el responsable de dicho programa.
Las solicitudes pueden enviarse hasta el 1 de julio de 2013.
- Listado provisional de admitidos y excluidos (subsanación de errores hasta el 13 de julio de 2013).
- Listado definitivo de admitidos y excluidos.
- Valoración provisional de méritos del concurso.
- Listado definitivo.
- Resolución definitiva.
Antimicrob Agents Chemother. 2013 Jun;57(6):2651-3
Rodríguez-Beltrán J, Rodríguez-Rojas A, Yubero E, Blázquez J.
 Animal fodder is routinely complemented with antibiotics together with other food supplements to improve growth. For instance, sepiolite is currently used as a dietary coadjuvant in animal feed, as it increases animal growth parameters and improves meat and derived final product quality. This type of food additive has so far been considered innocuous for the development and spread of antibiotic resistance.
Animal fodder is routinely complemented with antibiotics together with other food supplements to improve growth. For instance, sepiolite is currently used as a dietary coadjuvant in animal feed, as it increases animal growth parameters and improves meat and derived final product quality. This type of food additive has so far been considered innocuous for the development and spread of antibiotic resistance.
In this study, we demonstrate that sepiolite promotes the direct horizontal transfer of antibiotic resistance plasmids between bacterial species. The conditions needed for plasmid transfer (sepiolite and friction forces) occur in the digestive tracts of farm animals, which routinely receive sepiolite as a food additive. Furthermore, this effect may be aggravated by the use of antibiotics supplied as growth promoters.
Casado-Vela J, Matthiesen R, Sellés S, Naranjo JR.
 Understanding protein interaction networks and their dynamic changes is a major challenge in modern biology. Currently, several experimental and in silico approaches allow the screening of protein interactors in a large-scale manner. Therefore, the bulk of information on protein interactions deposited in databases and peer-reviewed published literature is constantly growing. Multiple databases interfaced from user-friendly web tools recently emerged to facilitate the task of protein interaction data retrieval and data integration. Nevertheless, as we evidence in this report, despite the current efforts towards data integration, the quality of the information on protein interactions retrieved by in silico approaches is frequently incomplete and may even list false interactions. Here we point to some obstacles precluding confident data integration, with special emphasis on protein interactions, which include gene acronym redundancies and protein synonyms.
Understanding protein interaction networks and their dynamic changes is a major challenge in modern biology. Currently, several experimental and in silico approaches allow the screening of protein interactors in a large-scale manner. Therefore, the bulk of information on protein interactions deposited in databases and peer-reviewed published literature is constantly growing. Multiple databases interfaced from user-friendly web tools recently emerged to facilitate the task of protein interaction data retrieval and data integration. Nevertheless, as we evidence in this report, despite the current efforts towards data integration, the quality of the information on protein interactions retrieved by in silico approaches is frequently incomplete and may even list false interactions. Here we point to some obstacles precluding confident data integration, with special emphasis on protein interactions, which include gene acronym redundancies and protein synonyms.
Three human proteins (choline kinase, PPIase and uromodulin) and three different web-based data search engines focused on protein interaction data retrieval (PSICQUIC, DASMI and BIPS) were used to explain the potential occurrence of undesired errors that should be considered by researchers in the field. We demonstrate that, despite the recent initiatives towards data standardization, manual curation of protein interaction networks based on literature searches are still required to remove potential false positives. A three-step workflow consisting of:
(i) data retrieval from multiple databases,
(ii) peer-reviewed literature searches, and
(iii) data curation and integration, is proposed as the best strategy to gather updated information on protein interactions.
Finally, this strategy was applied to compile bona fide information on human DREAM protein interactome, which constitutes liable training datasets that can be used to improve computational predictions.
J Immunol. 2013 Jun 15;190(12):6135-44
Fernández D, Ortiz M, Rodríguez L, García A, Martinez D, Moreno de Alborán I.
 The immune response involves the generation of Ab-secreting cells and memory B cells through a process called terminal B lymphocyte differentiation. This program requires the transcriptional repressor Blimp-1, which inhibits c-myc expression and terminates proliferation. Although the role of c-Myc in cell proliferation is well characterized, it is not known whether it has other functions in terminal differentiation.
The immune response involves the generation of Ab-secreting cells and memory B cells through a process called terminal B lymphocyte differentiation. This program requires the transcriptional repressor Blimp-1, which inhibits c-myc expression and terminates proliferation. Although the role of c-Myc in cell proliferation is well characterized, it is not known whether it has other functions in terminal differentiation.
In this study, we show that c-Myc not only regulates cell proliferation, but it is also essential for Ab-secreting cell function and differentiation in vivo. c-Myc–deficient B lymphocytes hypersecrete IgM and do not undergo Ig class switch recombination (CSR). CSR has been previously linked to proliferation, and in this study we mechanistically link class switching and proliferation via c-Myc. We observed that c-Myc regulates CSR by transcriptionally activating the B cell–specific factor activation-induced cytidine deaminase. By linking cell proliferation and CSR, c-Myc is thus a critical component for a potent immune response.
J Virol. 2013 Apr;87(8):4534-44
Alfonso R, Rodriguez A, Rodriguez P, Lutz T, Nieto A.
 The influenza virus polymerase associates to an important number of transcription-related proteins, including the largest subunit of the RNA polymerase II complex (RNAP II). Despite this association, degradation of the RNAP II takes place in the infected cells once viral transcription is completed. We have previously shown that the chromatin remodeler CHD6 protein interacts with the influenza virus polymerase complex, represses viral replication, and relocalizes to inactive chromatin during influenza virus infection.
The influenza virus polymerase associates to an important number of transcription-related proteins, including the largest subunit of the RNA polymerase II complex (RNAP II). Despite this association, degradation of the RNAP II takes place in the infected cells once viral transcription is completed. We have previously shown that the chromatin remodeler CHD6 protein interacts with the influenza virus polymerase complex, represses viral replication, and relocalizes to inactive chromatin during influenza virus infection.
In this paper, we report that CHD6 acts as a negative modulator of the influenza virus polymerase activity and is also subjected to degradation through a process that includes the following characteristics:
(i) the cellular proteasome is not implicated,
(ii) the sole expression of the three viral polymerase subunits from its cloned cDNAs is sufficient to induce proteolysis, and
(iii) degradation is also observed in vivo in lungs of infected mice and correlates with the increase of viral titers in the lungs.
Collectively, the data indicate that CHD6 degradation is a general effect exerted by influenza A viruses and suggest that this viral repressor may play an important inhibitory role since degradation and accumulation into inactive chromatin occur during the infection.
Cuellar J, Perales-Calvo J, Muga A, Valpuesta JM, Moro F.
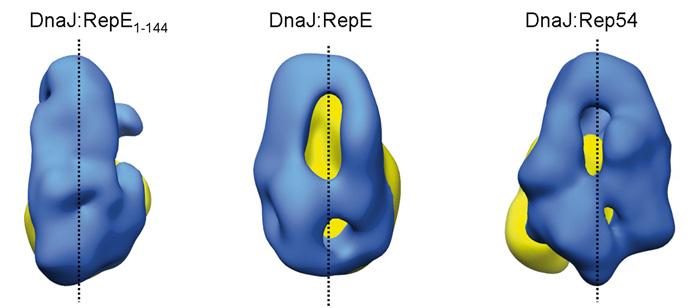 Hsp40 chaperones bind and transfer substrate proteins to Hsp70s and regulate their ATPase activity. The interaction of Hsp40s with native proteins modifies their structure and function. A good model for this function is DnaJ, the bacterial Hsp40 that interacts with RepE, the repressor/activator of plasmid F replication, and together with DnaK regulates its function. We characterize here the structure of the DnaJ:RepE complex by electron microscopy, the first described structure of a complex between an Hsp40 and a client protein. The comparison of the complexes of DnaJ with two RepE mutants reveals an intrinsic plasticity of the DnaJ dimer that allows the chaperone to adapt to different substrates. We also show that DnaJ induces conformational changes in dimeric RepE, which increase the intermonomeric distance and remodel both RepE domains enhancing its affinity for DNA.
Hsp40 chaperones bind and transfer substrate proteins to Hsp70s and regulate their ATPase activity. The interaction of Hsp40s with native proteins modifies their structure and function. A good model for this function is DnaJ, the bacterial Hsp40 that interacts with RepE, the repressor/activator of plasmid F replication, and together with DnaK regulates its function. We characterize here the structure of the DnaJ:RepE complex by electron microscopy, the first described structure of a complex between an Hsp40 and a client protein. The comparison of the complexes of DnaJ with two RepE mutants reveals an intrinsic plasticity of the DnaJ dimer that allows the chaperone to adapt to different substrates. We also show that DnaJ induces conformational changes in dimeric RepE, which increase the intermonomeric distance and remodel both RepE domains enhancing its affinity for DNA.
COOKIES POLICY
A cookie is a text file that is stored on your computer or mobile device via a web server and only that server will be able to retrieve or read the contents of the cookie and allow the Web site remember browser preferences and navigate efficiently. Cookies make the interaction between the user and the website faster and easier.
General information
This Website uses cookies. Cookies are small text files generated by the web pages you visit, which contain the session data that can be useful later in the website. In this way this Web remembers information about your visit, which can facilitate your next visit and make the website more useful.
How do cookies?
Cookies can only store text, usually always anonymous and encrypted. No personal information is ever stored in a cookie, or can be associated with identified or identifiable person.
The data allow this website to keep your information between the pages, and also to discuss how to interact with the website. Cookies are safe because they can only store information that is put there by the browser, which is information the user entered in the browser or included in the page request. You can not run the code and can not be used to access your computer. If a website encrypts cookie data, only the website can read the information.
What types of cookies used?
The cookies used by this website can be distinguished by the following criteria:
1. Types of cookies as the entity that manages:
Depending on who the entity operating the computer or domain where cookies are sent and treat the data obtained, we can distinguish:
- Own cookies: are those that are sent to the user's terminal equipment from a computer or domain managed by the editor itself and from which provides the service requested by the user.
- Third party cookies: these are those that are sent to the user's terminal equipment from a machine or domain that is not managed by the publisher, but by another entity data is obtained through cookies.
In the event that the cookies are installed from a computer or domain managed by the editor itself but the information collected by these is managed by a third party can not be considered as party cookies.
2. Types of cookies as the length of time that remain active:
Depending on the length of time that remain active in the terminal equipment can be distinguished:
- Session cookies: cookies are a type designed to collect and store data while the user accesses a web page. Are usually used to store information that only worth preserving for the service requested by the user at any one time (eg a list of products purchased).
- Persistent cookies: cookies are a type of data which are stored in the terminal and can be accessed and treated for a period defined by the head of the cookie, and can range from a few minutes to several years.
3. Cookies types according to their purpose:
Depending on the purpose for which the data are processed through cookies, we can distinguish between:
- Technical cookies: these are those that allow the user to navigate through a web page or application platform and the use of different options or services it exist as, for example, control traffic and data communication, identify the session, access to restricted access parts, remember the elements of an order, make the buying process an order, make an application for registration or participation in an event, use security features while browsing store content for dissemination videos or sound or share content via social networks.
- Customization cookies: these are those that allow the user to access the service with some general characteristics based on a predefined set of criteria in the user terminal would eg language, the type of browser through which you access the service, the locale from which you access the service, etc.
- Analysis cookies: they are those that allow the responsible for them, monitoring and analyzing the behavior of users of the web sites that are linked. The information gathered through such cookies are used in measuring the activity of web sites, application or platform and for the profiling of user navigation of such sites, applications and platforms, in order to make improvements function data analysis how users use the service.
Management tool cookies
This Website uses Google Analytics.
Google Analytics is a free tool from Google that primarily allows website owners know how users interact with your website. Also, enable cookies in the domain of the site in which you are and uses a set of cookies called "__utma" and "__utmz" to collect information anonymously and reporting of website trends without identifying individual users..
For statistics of use of this website use cookies in order to know the level of recurrence of our visitors and more interesting content. This way we can concentrate our efforts on improving the most visited areas and make the user more easily find what they are looking for. On this site you can use the information from your visit for statistical evaluations and calculations anonymous data and to ensure the continuity of service or to make improvements to their websites. For more details, see the link below privacy policy [http://www.google.com/intl/en/policies/privacy/]
How to manage cookies on your computer: disabling and deleting cookies
All Internet browsers allow you to limit the behavior of a cookie or disable cookies within settings or browser settings. The steps for doing so are different for each browser, you can find instructions in the help menu of your browser.
If you decline the use of cookies, since it is possible thanks to the preferences menu of your browser or settings, reject, this website will continue to function properly without the use of the same.
Can you allow, block or delete cookies installed on your computer by setting your browser options installed on your computer:
- For more information about Internet Explorer click here.
- For more information on Chrome click here.
- For more information about Safari click here.
- For more information about Firefox click here.
Through your browser, you can also view the cookies that are on your computer, and delete them as you see fit. Cookies are text files, you can open and read the contents. The data within them is almost always encrypted with a numeric key corresponding to an Internet session so often has no meaning beyond the website who wrote it.
Informed consent
The use of this website on the other hand, implies that you paid your specific consent to the use of cookies, on the terms and conditions provided in this Cookies Policy, without prejudice to the measures of deactivation and removal of cookies that you can take, and mentioned in the previous section.





