Según se van sintetizando las proteínas en los ribosomas, éstas tienen que ir tomando la forma adecuada para poder ejercer su función. Aunque las características físico-químicas de los aminoácidos que las forman determinan en gran parte la forma que adquieren, hay muchas proteínas que necesitan ayuda extra de parte de un grupo de proteínas conocidas como chaperonas.
 Para comprender mejor cómo funcionan estas proteínas, en su laboratorio del Centro Nacional de Biotecnología del CSIC (CNB), el grupo dirigido por José María Valpuesta ha utilizado la microscopía electrónica. Gracias a esta técnica han podido determinar por primera vez la estructura de un complejo formado por la chaperona DnaJ y su sustrato, lo que les ha permitido observar cómo la chaperona cambia la estructura del sustrato y con ello su función.
Para comprender mejor cómo funcionan estas proteínas, en su laboratorio del Centro Nacional de Biotecnología del CSIC (CNB), el grupo dirigido por José María Valpuesta ha utilizado la microscopía electrónica. Gracias a esta técnica han podido determinar por primera vez la estructura de un complejo formado por la chaperona DnaJ y su sustrato, lo que les ha permitido observar cómo la chaperona cambia la estructura del sustrato y con ello su función.
En colaboración con la Universidad del País Vasco, el investigador postdoctoral del CNB Jorge Cuéllar ha identificado además en dicha chaperona una zona de gran flexibilidad que le permite adaptarse a la forma de distintas proteínas. Como se puede apreciar en la imagen de abajo, la forma que adopta la chaperona DnaJ (en azul) cambia radicalmente en función del sustrato al que se une (RepE1-144, RepE o Rep54; en amarillo). De este modo, una misma chaperona es capaz de unirse a una variedad de proteínas diferentes, consiguiendo en todas ellas que adquieran la forma necesaria para funcionar.
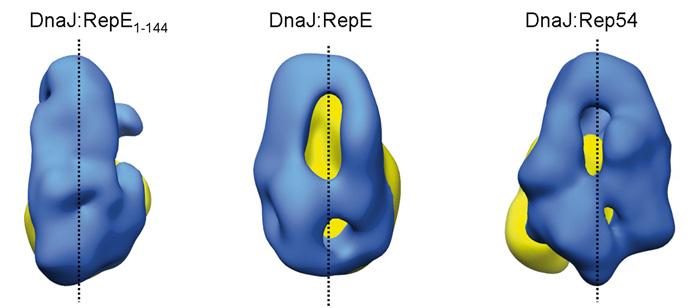
- Cuéllar J, Perales-Calvo J, Muga A, Valpuesta JM, Moro F. Structural insights into the chaperone activity of the 40 kDa heat shock protein DnaJ. Binding and remodeling of a native substrate. J Biol Chem 2013 Apr 11. [doi: 10.1074/jbc.M112.430595].
Nucleic Acids Res. 2013 Feb 1;41(3):1711-21
Seco EM, Zinder JC, Manhart CM, Lo Piano A, McHenry CS, Ayora S.
 Complex viruses that encode their own initiation proteins and subvert the host's elongation apparatus have provided valuable insights into DNA replication. Using purified bacteriophage SPP1 and Bacillus subtilis proteins, we have reconstituted a rolling circle replication system that recapitulates genetically defined protein requirements.
Complex viruses that encode their own initiation proteins and subvert the host's elongation apparatus have provided valuable insights into DNA replication. Using purified bacteriophage SPP1 and Bacillus subtilis proteins, we have reconstituted a rolling circle replication system that recapitulates genetically defined protein requirements.
Eleven proteins are required: phage-encoded helicase (G40P), helicase loader (G39P), origin binding protein (G38P) and G36P single-stranded DNA-binding protein (SSB); and host-encoded PolC and DnaE polymerases, processivity factor (β(2)), clamp loader (τ-δ-δ') and primase (DnaG). This study revealed a new role for the SPP1 origin binding protein. In the presence of SSB, it is required for initiation on replication forks that lack origin sequences, mimicking the activity of the PriA replication restart protein in bacteria. The SPP1 replisome is supported by both host and viral SSBs, but phage SSB is unable to support B. subtilis replication, likely owing to its inability to stimulate the PolC holoenzyme in the B. subtilis context. Moreover, phage SSB inhibits host replication, defining a new mechanism by which bacterial replication could be regulated by a viral factor.
Sánchez-Sampedro L, Gómez CE, Mejías-Pérez E, Pérez-Jiménez E, Oliveros JC, Esteban M.
 Replication competent poxvirus vectors with an attenuation phenotype and with high immunogenic capacity of the foreign expressed antigen are being pursuit as novel vaccine vectors against different pathogens. In this investigation we have examined the replication and immunogenic characteristics of two vaccinia virus (VACV) mutants M65 and M101.
Replication competent poxvirus vectors with an attenuation phenotype and with high immunogenic capacity of the foreign expressed antigen are being pursuit as novel vaccine vectors against different pathogens. In this investigation we have examined the replication and immunogenic characteristics of two vaccinia virus (VACV) mutants M65 and M101.
These mutants were generated after 65 and 101 serial passages of persistently infected Friend erythroleukemia cells (FEL). In cultured cells of different origins the mutants are replication competent and have growth kinetics similar or slightly reduced in comparison with the parental Western Reserve (WR) virus strain. In normal and immune suppressed infected mice the mutants showed different attenuation levels, and pathogenicity in comparison with WR and MVA strains. Wide genome analysis after deep sequencing revealed selected genomic deletions and mutations in a number of viral open-reading frames (ORFs). Mice immunized in DNA prime/Mutant boost regime with viral vectors expressing the LACK antigen of Leismania infantum resulted in protection or delay in the onset of cutaneus leishmaniasis. Protection was similar to that triggered by MVA-LACK. In immunized mice, both polyfunctional CD4+ and CD8+ T cells with an effector memory phenotype were activated by the two mutants, but DNA-LACK/M65-LACK protocol preferentially induced CD4+ while DNA-LACK/M101-LACK preferentially induced CD8+ T cell responses.
Altogether our findings showed the adaptive changes of the WR genome during long-term virus-host cell interaction and how replication competency of M65 and M101 mutants confers distinct biological properties and immunogenicity in mice, as compared to the MVA strain. These mutants could have applicability for understanding VACV biology and as potential vaccine vectors against pathogens and tumors.
J Virol. 2013 Jul;87(13):7282-300
Gómez CE, Perdiguero B, Cepeda MV, Mingorance L, García-Arriaza J, Vandermeeren A, Sorzano CO, Esteban M.
 A major goal in the control of hepatitis C infection is the development of a vaccine. Here, we have developed a novel HCV vaccine candidate based on the highly attenuated poxvirus vector MVA (referred as MVA-HCV) expressing the near full-length (7.9 kbp) HCV sequence, with the aim to target almost all of the T and B cell determinants described for HCV.
A major goal in the control of hepatitis C infection is the development of a vaccine. Here, we have developed a novel HCV vaccine candidate based on the highly attenuated poxvirus vector MVA (referred as MVA-HCV) expressing the near full-length (7.9 kbp) HCV sequence, with the aim to target almost all of the T and B cell determinants described for HCV.
In infected cells, MVA-HCV produces a polyprotein that is subsequently processed into the structural and nonstructural HCV proteins, triggering the cytoplasmic accumulation of dense membrane aggregates. In both C57BL/6 and transgenic HLA-A2 vaccinated mice, MVA-HCV induced high, broad, polyfunctional and long-lasting HCV-specific T cell immune responses. The vaccine-induced T cell response was mainly mediated by CD8 T cells; however, although lower in magnitude, the CD4+ T cells were highly polyfunctional. In homologous protocol (MVA-HCV/MVA-HCV) the main CD8+ T cell target was p7+NS2, whereas in heterologous combination (DNA-HCV/MVA-HCV) the main target was NS3. Antigenic responses were also detected against other HCV proteins (Core, E1-E2, NS4) but the magnitude of the responses was dependent of the protocol used. The majority of the HCV-induced CD8+ T cells were triple or quadruple-cytokine producers. The MVA-HCV vaccine induced memory CD8+ T cell responses with an effector memory (TEM) phenotype.
Overall, our data showed that MVA-HCV induced broad, highly polyfunctional and durable T cell responses of a magnitude and quality that might be associated with protective immunity and open the path for future considerations of MVA-HCV as prophylactic and/or therapeutic vaccine candidate against HCV.
Un estudio internacional en el que ha participado la investigadora del CNB Cristina Risco ha publicado en la revista PNAS la estructura de la ribonucleoproteína del virus Bunyamwera, modelo de estudio de los integrantes de la familia Bunyaviridae, a la que también pertenecen los virus que provocan la fiebre hemorrágica de Congo y Crimea y la fiebre del valle del Rift.
"Uno de los principales resultados de esta investigación es que la flexibilidad de la nucleoproteína que envuelve el ARN de Bunyamwera facilita el empaquetamiento de su material genético en las partículas virales y su replicación una vez fuera del virus. La estructura atómica muestra que el plegamiento de la nucleoproteína es, además, diferente al observado en las demás nucleoproteínas de virus conocidas”, explica Risco.
Este estudio muestra que, a pesar de su flexibilidad, el complejo que recubre el material genético lo protege completamente del ataque de las nucleasas, las enzimas que rompen las moléculas de ARN.
Los resultados de este estudio ayudan a entender mejor el mecanismo molecular mediante el que se ensamblan las ribonucleoproteínas virales. "Este conocimiento permitirá avanzar en el desarrollo de compuestos para atacar a estos complejos macromoleculares que son esenciales para la supervivencia de los virus ARN", añade desde el mismo laboratorio del Centro Nacional de Biotecnología del CSIC la investigadora Isabel Fernández de Castro.
Los hallazgos logrados por este estudio, liderado por investigadores chinos, ha sido posible mediante el uso de cristalografía de rayos X y microscopía electrónica. Como se aprecia en la imagen, el complejo formado por el monómero de la proteína viral NP y el ARN, la flexibilidad de este complejo respecto a los adyacentes es el elemento básico que dirige la oligomerización y ensamblaje de las ribonucleoproteínas lineales y flexibles del virus Bunyamwera.
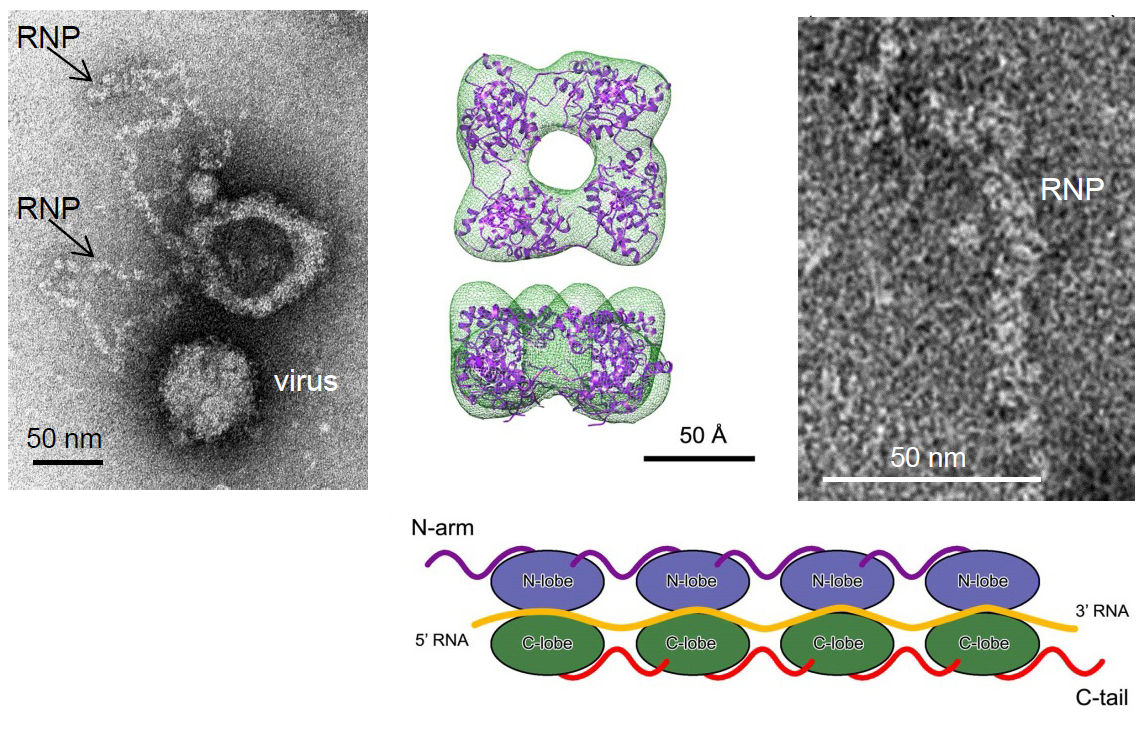
- Li B, Wang Q, Pan X, Fernández de Castro I, Sun Y, Guo Y, Tao X, Risco C, Su SF, Lou Z. Bunyamwera virus possesses a distinct nucleocapsid protein to facilitate genome encapsidation. PNAS April 8, 2013 doi: 10.1073/pnas.1222552110.
Li B, Wang Q, Pan X, Fernández de Castro I, Sun Y, Guo Y, Tao X, Risco C, Su SF, Lou Z.
 Bunyamwera virus (BUNV), which belongs to the genus Orthobunyavirus, is the prototypical virus of the Bunyaviridae family. Similar to other negative-sense single-stranded RNA viruses, bunyaviruses possess a nucleocapsid protein (NP) to facilitate genomic RNA encapsidation and virus replication.
Bunyamwera virus (BUNV), which belongs to the genus Orthobunyavirus, is the prototypical virus of the Bunyaviridae family. Similar to other negative-sense single-stranded RNA viruses, bunyaviruses possess a nucleocapsid protein (NP) to facilitate genomic RNA encapsidation and virus replication.
The structures of two NPs of members of different genera within the Bunyaviridae family have been reported. However, their structures, RNA-binding features, and functions beyond RNA binding significantly differ from one another. Here, we report the crystal structure of the BUNV NP–RNA complex. The polypeptide of the BUNV NP was found to possess a distinct fold among viral NPs. An N-terminal arm and a C-terminal tail were found to interact with neighboring NP protomers to form a tetrameric ring-shaped organization. Each protomer bound a 10-nt RNA molecule, which was acquired from the expression host, in the positively charged crevice between the N and C lobes. Inhomogeneous oligomerization was observed for the recombinant BUNV NP–RNA complex, which was similar to the Rift Valley fever virus NP–RNA complex.
This result suggested that the flexibility of one NP protomer with adjacent protomers underlies the BUNV ribonucleoprotein complex (RNP) formation. Electron microscopy revealed that the monomer-sized NP–RNA complex was the building block of the natural BUNV RNP. Combined with previous results indicating that mutagenesis of the interprotomer or protein–RNA interface affects BUNV replication, our structure provides a great potential for understanding the mechanism underlying negative-sense single-stranded RNA RNP formation and enables the development of antiviral therapies targeting BUNV RNP formation.
Castón JR, Luque D, Gómez-Blanco J, Ghabrial SA.
 Chrysoviruses are double-stranded RNA viruses with a multipartite genome. Structure of two fungal chrysoviruses, Penicillium chrysogenum virus and Cryphonectria nitschkei chrysovirus 1, has been determined by three-dimensional cryo-electron microscopy analysis and in hydrodynamic studies.
Chrysoviruses are double-stranded RNA viruses with a multipartite genome. Structure of two fungal chrysoviruses, Penicillium chrysogenum virus and Cryphonectria nitschkei chrysovirus 1, has been determined by three-dimensional cryo-electron microscopy analysis and in hydrodynamic studies.
The capsids of both viruses are based on a T=1 lattice containing 60 subunits, remain structurally undisturbed throughout the viral cycle, and participate in genome metabolism. The capsid protein is formed by a repeated α-helical core, indicative of gene duplication.
Whereas the chrysovirus capsid protein has two motifs with the same fold, most dsRNA virus capsid subunits consist of dimers of a single protein with similar folds. The arrangement of the chrysovirus α-helical core is conserved in the totivirus L-A capsid protein, suggesting a shared basic fold. The encapsidated genome is organized in concentric shells; whereas inner dsRNA shells are diffuse, the outermost layer is organized into a dodecahedral cage beneath the protein capsid. This genome ordering could constitute a framework for dsRNA transcription in the capsid interior and/or have a structural role for capsid stability.
El Campus de Excelencia Internacional UAM+CSIC (CEI UAM+CSIC) tiene como una de sus líneas de actuación esenciales la captación de talento tanto a nivel nacional como internacional. Y con la intención de atraer científicos y profesores de primer nivel, acaba de convocar el Premio de Investigación CEI UAM+CSIC en el área de la Biología Molecular Física y Sintética.
 El objetivo de este premio, compatible con cualquier otro tipo de premio y/o ayuda, es el de favorecer que profesionales de otros centros de investigación puedan trasladarse al CNB para desarrollar una línea de investigación que se centre en el estudio del plegamiento y función de proteínas. Para ello, deberán integrar métodos experimentales de alta resolución temporal, estructural y de moléculas únicas, modelización teórica y simulaciones computacionales.
El objetivo de este premio, compatible con cualquier otro tipo de premio y/o ayuda, es el de favorecer que profesionales de otros centros de investigación puedan trasladarse al CNB para desarrollar una línea de investigación que se centre en el estudio del plegamiento y función de proteínas. Para ello, deberán integrar métodos experimentales de alta resolución temporal, estructural y de moléculas únicas, modelización teórica y simulaciones computacionales.
El premio tiene una dotación económica de 30.000 € anuales de libre disposición durante tres años, prorrogables a otros dos. Para solicitarlo, el investigador debe tener una vinculación permanente con el CSIC y no podrá estar adscrito a ningún centro del CEI UAM+CSIC.
Las solicitudes se presentarán en el registro general de la UAM desde el 8 de abril al 22 de abril de 2013.
Philos Trans R Soc Lond B Biol Sci. 2013 Mar 11;368(1616):20120377
Nikel PI, Pérez-Pantoja D, de Lorenzo V.
 Chlorinated pollutants are hardly biodegradable under oxic conditions, but they can often be metabolized by anaerobic bacteria through organohalide respiration reactions.
Chlorinated pollutants are hardly biodegradable under oxic conditions, but they can often be metabolized by anaerobic bacteria through organohalide respiration reactions.
In an attempt to identify bottlenecks limiting aerobic catabolism of 1,3-dichloroprop-1-ene (1,3-DCP; a widely used organohalide) in Pseudomonas pavonaceae, the possible physiological restrictions for this process were surveyed. Flow cytometry and a bioluminescence reporter of metabolic state revealed that cells treated with 1,3-DCP experienced an intense stress that could be traced to the endogenous production of reactive oxygen species (ROS) during the metabolism of the compound. Cells exposed to 1,3-DCP also manifested increased levels of d-glucose-6-P 1-dehydrogenase activity (G6PDH, an enzyme key to the synthesis of reduced NADPH), observed under both glycolytic and gluconeogenic growth regimes. The increase in G6PDH activity, as well as cellular hydroperoxide levels, correlated with the generation of ROS. Additionally, the high G6PDH activity was paralleled by the accumulation of d-glucose-6-P, suggesting a metabolic flux shift that favours the production of NADPH. Thus, G6PDH and its cognate substrate seem to play an important role in P. pavonaceae under redox stress caused by 1,3-DCP, probably by increasing the rate of NADPH turnover.
The data suggest that oxidative stress associated with the biodegradation of 1,3-DCP reflects a significant barrier for the evolution of aerobic pathways for chlorinated compounds, thereby allowing for the emergence of anaerobic counterparts.
In this paper we have addressed the problem of analysing Next Generation Sequencing samples with an expected large biodiversity content. We analysed several well-known 16S rRNA datasets from experimental samples, including both large and short sequences, in numbers of tens of thousands, in addition to carefully crafted synthetic datasets containing more than 7000 OTUs.
From this data analysis several patterns were identified and used to develop new guidelines for experimentation in conditions of high biodiversity. We analysed the suitability of different clustering packages for these type of situations, the problem of even sampling, the relative effectiveness of Chao1 and ACE estimators as well as their effect on sampling size for a variety of population distributions. As regards practical analysis procedures, we advocated an approach that retains as much high-quality experimental data as possible. By carefully applying selection rules combining the taxonomic assignment with clustering strategies, we derived a set of recommendations for ultra-sequencing data analysis at high biodiversity levels.
COOKIES POLICY
A cookie is a text file that is stored on your computer or mobile device via a web server and only that server will be able to retrieve or read the contents of the cookie and allow the Web site remember browser preferences and navigate efficiently. Cookies make the interaction between the user and the website faster and easier.
General information
This Website uses cookies. Cookies are small text files generated by the web pages you visit, which contain the session data that can be useful later in the website. In this way this Web remembers information about your visit, which can facilitate your next visit and make the website more useful.
How do cookies?
Cookies can only store text, usually always anonymous and encrypted. No personal information is ever stored in a cookie, or can be associated with identified or identifiable person.
The data allow this website to keep your information between the pages, and also to discuss how to interact with the website. Cookies are safe because they can only store information that is put there by the browser, which is information the user entered in the browser or included in the page request. You can not run the code and can not be used to access your computer. If a website encrypts cookie data, only the website can read the information.
What types of cookies used?
The cookies used by this website can be distinguished by the following criteria:
1. Types of cookies as the entity that manages:
Depending on who the entity operating the computer or domain where cookies are sent and treat the data obtained, we can distinguish:
- Own cookies: are those that are sent to the user's terminal equipment from a computer or domain managed by the editor itself and from which provides the service requested by the user.
- Third party cookies: these are those that are sent to the user's terminal equipment from a machine or domain that is not managed by the publisher, but by another entity data is obtained through cookies.
In the event that the cookies are installed from a computer or domain managed by the editor itself but the information collected by these is managed by a third party can not be considered as party cookies.
2. Types of cookies as the length of time that remain active:
Depending on the length of time that remain active in the terminal equipment can be distinguished:
- Session cookies: cookies are a type designed to collect and store data while the user accesses a web page. Are usually used to store information that only worth preserving for the service requested by the user at any one time (eg a list of products purchased).
- Persistent cookies: cookies are a type of data which are stored in the terminal and can be accessed and treated for a period defined by the head of the cookie, and can range from a few minutes to several years.
3. Cookies types according to their purpose:
Depending on the purpose for which the data are processed through cookies, we can distinguish between:
- Technical cookies: these are those that allow the user to navigate through a web page or application platform and the use of different options or services it exist as, for example, control traffic and data communication, identify the session, access to restricted access parts, remember the elements of an order, make the buying process an order, make an application for registration or participation in an event, use security features while browsing store content for dissemination videos or sound or share content via social networks.
- Customization cookies: these are those that allow the user to access the service with some general characteristics based on a predefined set of criteria in the user terminal would eg language, the type of browser through which you access the service, the locale from which you access the service, etc.
- Analysis cookies: they are those that allow the responsible for them, monitoring and analyzing the behavior of users of the web sites that are linked. The information gathered through such cookies are used in measuring the activity of web sites, application or platform and for the profiling of user navigation of such sites, applications and platforms, in order to make improvements function data analysis how users use the service.
Management tool cookies
This Website uses Google Analytics.
Google Analytics is a free tool from Google that primarily allows website owners know how users interact with your website. Also, enable cookies in the domain of the site in which you are and uses a set of cookies called "__utma" and "__utmz" to collect information anonymously and reporting of website trends without identifying individual users..
For statistics of use of this website use cookies in order to know the level of recurrence of our visitors and more interesting content. This way we can concentrate our efforts on improving the most visited areas and make the user more easily find what they are looking for. On this site you can use the information from your visit for statistical evaluations and calculations anonymous data and to ensure the continuity of service or to make improvements to their websites. For more details, see the link below privacy policy [http://www.google.com/intl/en/policies/privacy/]
How to manage cookies on your computer: disabling and deleting cookies
All Internet browsers allow you to limit the behavior of a cookie or disable cookies within settings or browser settings. The steps for doing so are different for each browser, you can find instructions in the help menu of your browser.
If you decline the use of cookies, since it is possible thanks to the preferences menu of your browser or settings, reject, this website will continue to function properly without the use of the same.
Can you allow, block or delete cookies installed on your computer by setting your browser options installed on your computer:
- For more information about Internet Explorer click here.
- For more information on Chrome click here.
- For more information about Safari click here.
- For more information about Firefox click here.
Through your browser, you can also view the cookies that are on your computer, and delete them as you see fit. Cookies are text files, you can open and read the contents. The data within them is almost always encrypted with a numeric key corresponding to an Internet session so often has no meaning beyond the website who wrote it.
Informed consent
The use of this website on the other hand, implies that you paid your specific consent to the use of cookies, on the terms and conditions provided in this Cookies Policy, without prejudice to the measures of deactivation and removal of cookies that you can take, and mentioned in the previous section.





