Hernando-Pérez M, Martín-González N, Pérez-Illana M, Suomalainen M, Condezo GN, Philomena Ostapchuk P, Gallardo J, Menéndez M, Greber UF, Hearing P, de Pablo PJ, San Martín C.
Abstract
Adenovirus minor coat protein VI contains a membrane-disrupting peptide that is inactive when VI is bound to hexon trimers. Protein VI must be released during entry to ensure endosome escape. Hexon:VI stoichiometry has been uncertain, and only fragments of VI have been identified in the virion structure. Recent findings suggest an unexpected relationship between VI and the major core protein, VII. According to the high-resolution structure of the mature virion, VI and VII may compete for the same binding site in hexon; and noninfectious human adenovirus type 5 particles assembled in the absence of VII (Ad5-VII-) are deficient in proteolytic maturation of protein VI and endosome escape. Here we show that Ad5-VII- particles are trapped in the endosome because they fail to increase VI exposure during entry. This failure was not due to increased particle stability, because capsid disruption happened at lower thermal or mechanical stress in Ad5-VII- compared to wild-type (Ad5-wt) particles. Cryoelectron microscopy difference maps indicated that VII can occupy the same binding pocket as VI in all hexon monomers, strongly arguing for binding competition. In the Ad5-VII- map, density corresponding to the immature amino-terminal region of VI indicates that in the absence of VII the lytic peptide is trapped inside the hexon cavity, and clarifies the hexon:VI stoichiometry conundrum. We propose a model where dynamic competition between proteins VI and VII for hexon binding facilitates the complete maturation of VI, and is responsible for releasing the lytic protein from the hexon cavity during entry and stepwise uncoating.
- Los dos proyectos que se desarrollan en el CNB están progresando muy rápido, todo lo técnicamente posible, y cuentan con los recursos necesarios gracias a la financiación del Gobierno
- El proyecto de Enjuanes y Sola ha conseguido un ADN complementario completo del coronavirus SARS-CoV-2 y en paralelo se han preparado los test en modelos animales
- El proyecto de Mariano Esteban cuenta ya con un candidato a vacuna y se han vacunado ratones para probar la respuesta inmune protectiva frente al coronavirus
NEW: 23rd JUNE Provisional list of applicants admitted and excluded
NEW: 9th JULY: Definitive list of applicants admitted and excluded
NEW: 22 JULY: ALLOCATIONS
Submissions are open for the new JAE Intro SOMdM 2020 Scholarships at CSIC Severo Ochoa Centres of Excellence and María de Maeztu Units of Excellence.
This call is open to students who are going to enroll in a University Master's Degree during the 2020/2021 academic year. Applicants must have completed a degree in the academic year 2018/2019 or later (check the exceptions in the Call bases) and credit an average grade equal to or greater than 8.00. Applications deadline is June 15, 2020.
Call bases can be found here
In this call, CNB-CSIC, offers up to 6 scholarships (one per department), and each one will be allocated with 7200 euros in nine monthly installments. For more information on the groups offered, check our list of departments.
The Spanish National Centre for Biotechnology is a multidisciplinary research center with a long-standing experience in molecular and structural virology and immunology. To offer responses to the current COVID19 pandemic, CNB-CSIC has undertaken a series os collaborative and interdisciplinary studies that exploit synergies between research groups and scientific services.
Our lines of action comprise more than fifteen projects led by researchers from the centre. These projects are included in the CSIC Global Health Platform, a large inititive that involves over 200 research groups approaching the scientific challenges caused by COVID-19 pandemic to provide short, medium and long-term solutions.
 Development of a SARS-CoV2 vaccine based in non-infective replicons
Development of a SARS-CoV2 vaccine based in non-infective replicons
Researchers: Luis Enjuanes, Isabel Sola, Sonia Zuñiga
The aim of this project is to generate the SARS-CoV-2 virus in the laboratory by assembling synthetic DNA fragments and removing the genes responsible for virulence from its genome with reverse genetics techniques to introduce attenuating mutations and obtain non-infective and highly immunogenic derivatives as SARS-CoV-2 vaccine candidates. In parallel, animal models (transgenic mice) are being developed for the validation of vaccines and other therapeutic agents against COVID-19.
 Development of a SARS-CoV2 vaccine based in a non-replicative MVA vector containing specific viral antigens
Development of a SARS-CoV2 vaccine based in a non-replicative MVA vector containing specific viral antigens
Researchers: Mariano Esteban, Juan García Arriaza, Carmen E. Gómez
The aim is to generate viral vectors based on a modification of the Vaccinia virus (called MVA) containing some of the proteins on the surface of the virus that are relevant to the infection. Because of its proven high attenuation, as MVA recombinants have been tested in numerous clinical trials and the MVA vector has been licensed by the FDA and EMA regulatory agencies as a smallpox vaccine, the MVA-COVID-19 vaccine would be safe and could be administered at all ages to all types of populations, including people with immune deficiencies. Two different vaccine candidates will be initially tested in pre-clinical studies in mice, and the best vaccine candidate will be ready to enter clinical studies in humans.
 Identification and testing of antiviral compounds for the treatment of SARS-CoV2
Identification and testing of antiviral compounds for the treatment of SARS-CoV2
Researchers: Pablo Gastaminza, Urtzi Garaigorta
Platform for the massive screening of antiviral compounds, by means of two phases, one that is developed in the P2 laboratory with a model of the common cold coronavirus and another that is carried out in the P3 laboratory where the selected compounds are evaluated against SARS2. Repositioning drugs that are already in clinical use and would therefore be a quick way to find effective treatments in humans are being studied, as well as libraries of compounds, in collaboration with other projects led by CSIC groups, to identify new compounds and analyse their antiviral action mechanism. In collaboration with other projects led by CSIC groups, the screening of different collections of molecules is being carried out, which is making it possible to address both the repositioning of compounds for clinical use and the exploration of new antiviral compounds.
 Preclinical validation of therapeutical agents for SARS-CoV2 treatment
Preclinical validation of therapeutical agents for SARS-CoV2 treatment
Researchers: Luis Enjuanes, Isabel Sola, Sonia Zuñiga, Leonor Kremer
This research group has different in vitro and in vivo models for the study of human coronavirus pathogenesis and is finalizing the development of specific models for SARS-CoV-2. These models will be used for preclinical validation of therapeutic compounds against SARS-CoV-2, including own developments and projects in collaboration with the Spanish pharmaceutical industry for drug repositioning and immunotherapy strategies.
 Computational models for the design of therapeutic interventions against SARS-CoV-2
Computational models for the design of therapeutic interventions against SARS-CoV-2
Researchers: José Ramón Valverde, Luis Enjuanes, J. Manuel Honrubia
This line of action applies computational methods to several key aspects of SARS-CoV-2 research, including the modelling of mechanisms by which the virus could trigger the different symptoms of COVID-19, the analysis of antibody interaction with SARS-CoV-2 for the design of neutralising antibodies, as well as in silico studies of the dynamics of interaction between surface proteins of the virus and its receptor in order to generate a humanised mouse model for pre-clinical studies.
 Development of nanobodies against SARS-CoV2
Development of nanobodies against SARS-CoV2
Researchers: Luis Ángel Fernández, José María Casasnovas, Víctor de Lorenzo
The aim of this line of action is to obtain neutralising antibodies against SARS-CoV-2, using the technology of generating single domain antibodies with a human VH region and a camelid VHH domain (nanobodies) in bacteria, starting from available nanobody libraries, libraries that are being generated by dromedary immunisation with SARS-CoV-2 antigens, human antibody collections, or by synthesis and mutagenesis of neutralising antibodies.
 Design of immunosuppresive nanoparticles to fight the “cytokines storm” that causes lung immunopathology
Design of immunosuppresive nanoparticles to fight the “cytokines storm” that causes lung immunopathology
Researcher: Domingo F. Barber
Design of nanoparticles that can be used to reduce the inflammation that occurs, especially in the lungs, when infected with SARS-CoV-2. The project will make it possible to understand how iron oxide nanoparticles (NPOH), which are normally used in magnetic resonances or for the treatment of anaemia (FeraSpin™y Feraheme), interfere with the reproduction of the virus, as well as to design NPOHs that can be used to reduce the lung inflammation generated in some cases of SARS2 infection. These nanoparticles could also be used to mitigate the inflammation caused by a high number of inflammatory diseases.
 Use of CRISPR-Cas1 to destroy SARSCoV2 genome
Use of CRISPR-Cas1 to destroy SARSCoV2 genome
Researcher: Dolores Rodríguez
To use a new variant of the CRISPR gene editing tool and the Cas13d protein to destroy the SARS-CoV-2 coronavirus genome.The non-toxicity and efficiency of CRISPR reagents and Cas13d protein will be validated in vivo in zebrafish embryos. The efficiency of the project will be explored with the help of two related viruses (equine torovirus BEV, human coronavirus 229E) that allow working under BSL2 security conditions, before adapting the project to focus directly on the SARS-CoV-2 coronavirus genome under BSL3 conditions.
 Assays for the identification of individuals seropositive for COVID-19 using recombinant antigens produced in different expression systems (bacteria, insect or mammalian cells).
Assays for the identification of individuals seropositive for COVID-19 using recombinant antigens produced in different expression systems (bacteria, insect or mammalian cells).
Researchers: Francisco Rodríguez, Hugh Reyburn, José María Casasnovas, Mar Valés, José Miguel Rodríguez Frade
As a result of the collaboration between several research groups, it has been possible to develop the production of different SARS-CoV-2 proteins and their incorporation into diagnostic tests for seropositivity. The capacity of these antigens to detect the presence of antibodies against SARS-CoV-2 in serums of convalescent patients opens the way to the development of diagnostic tests to identify different stages of immunity in SARS-CoV-2 infected individuals, a key tool for epidemiological studies, to detect the presence of neutralizing antibodies or to temporarily monitor patients’ immunity. Very important aspects in the processes of de-escalation and return to the routine.
 Immunity response characterization to control the pandemic
Immunity response characterization to control the pandemic
Researchers: Margarita del Val (CBMSO) and Isabel Mérida
Short term [3-6 months]: to determine the % of a homogeneous population, in particular children between 5-16 years old that will be immunized against the coronavirus, which will allow to estimate the vulnerability of the Spanish population to a new wave of the COVID-19 infection. Medium term [1 year]: to know the capacity of the SARS-CoV-2 virus to produce the COVID-19 disease in the population and its associated pathologies. And, based on this, to guide therapies for its treatment and the development of vaccines. /p>
 Prediction of the COVID-19 epidemic dynamics
Prediction of the COVID-19 epidemic dynamics
Researchers: Susanna Manrubia, Saúl Ares
The focus of this project is the development of tools to manage, through predictive models, the appropriate social distancing measures to contain or prevent the expansion of COVID-19. The first phase of the project focuses on the analysis of data on the evolution of the pandemic in Spain and on the development of predictive and consensual models. In a second phase, the refinement of predictive models and potentially useful reports for a future pandemic and the study of explanatory models will be tackled.
 Development and experimental validation of sterilization and decontamination systems for the inactivation of SARS-CoV-2
Development and experimental validation of sterilization and decontamination systems for the inactivation of SARS-CoV-2
Researchers: Fernando Usera
The laboratory of biological containment level 3 (NCB-3) is the key infrastructure of the CNB for carrying out experiments with the SARS-CoV-2. In addition, the laboratory participates in a series of studies in collaboration with public entities and companies that aim to develop and validate sterilization and decontamination methods for the inactivation of SARS-CoV-2 in different environments and contaminated surfaces.
 SARS-CoV-2 Proteomics
SARS-CoV-2 Proteomics
Researchers: Fernando Corrales, Alberto Paradela
To provide the basis for new diagnostic, therapeutic and vaccination strategies needed to control the present and future pandemics. Short term: Development of therapeutic molecules by facilitating quality control methods. Medium term: To define the humoral immune response of patients with COVID-19 (our body’s protective response to extracellular particles of the virus). Diagnosis and stratification of patients. Long term: To define mechanisms associated with the infection process. New diagnostic and/or therapeutic routes.
 Structural biology of SARS-CoV-2
Structural biology of SARS-CoV-2
Researchers: José María Valpuesta, José Ruiz Castón, Jaime Martín Benito, Carmen San Martín, Mark van Raaij
CWith the goal of guiding the design of antibodies, antiviral compounds and other molecular tools to combat COVID-19, X-ray crystallography, electron cryomicroscopy and correlative cryomicroscopy techniques are being used to understand the structure of the virus, its components and the infection process. In addition, work is currently underway to design chimeric viral pseudo-particles from SARS-CoV-2 antigenic regions to validate their potential to generate a protective response to SARS-CoV-2.
 SARS-CoV-2 structure-function information node
SARS-CoV-2 structure-function information node
Researchers: José María Carazo, Carlos Óscar Sánchez-Sorzano
The aim of this project is to integrate and make available to the scientific community in the shortest time possible all the structural and biomedical information that is being generated in the world around SARS-CoV-2. In coordination with the Instituto Nacional de Bioinformática and the European infrastructures Instruct, ELIXIR y EOSC, a powerful interactive web information node called 3DBionotes is being built with the aim of being hosted on a future COVID-19 portal of the European Open Science Cloud (EOSC) initiative.
Due to the social impact of SARS-CoV2 pandemic, both CSIC and the CNB are receiving donations from differents institutions, private companies and individuals. We would like to express our gratitude to both companies and anonymous individuals for their involvement in our research.

Nueva convocatoria para solicitar becas JAE de Introducción a la Investigación en centros del CSIC. Las 250 becas ofertadas becas están orientadas a estudiantes universitarios, especialmente aquellos en el último curso de grado universitario y tendrán una duración de 5 meses consecutivos, iniciándose en septiembre u octubre de 2020.
El plazo de presentación de solicitudes se ha vuelto a abrir y termina el 10 de junio (incluido)
El Centro Nacional de Biotecnología recibirá 11 estudiantes en el curso 2020-2021
16 de JULIO 2020: RESOLUCION DEFINITIVA publicada en la página del CSIC
Next Wednesday 29th of January, The CNB will host a simposium to honour Amelia Nieto, a CNB researcher from the origins of the center.
For her retirement, several researchers who initiated their scientific career in her group are visiting the CNB to celebrate the meeting "Looking at Cell Biology from a virus perspective".
This event count with speakers such as Maite Huarte, Tomás Aragón and Puri Fortes from the Centro de Investigaciones Médicas Aplicadas (CIMA) din Pamplona; Juan Varcarcel and Susana de la Luna from the Centre Reserca Genòmica (CRG) in Barcelona; aSantiago F. González from the Università della Svizzera in Bellinzona and Emilio Yángüez from the Functional Genomic Service at ETH in Zurich will join the .
This meeting is supported by The Company of Biologist through their Small Meeting Grant program.
Attendance is free


- Carmen Elena Gómez receives the Research Prize from the National Royal Academy of Pharmacy (RANF)
- Mariano Esteban receives the Carracido Gold Medal for his scientific and academic career
Carmen Elena Gómez, researcher at the Molecular and Cellular Biology Department received (ex aequo) the Research Prize from the Real Academia Nacional de Farmacia (RANF). The award ceremony took place in the Academia the 15th of January 2020. 
This award recognises the research work “MVA-B as a model vaccine candidate against VIH/SIDA: From basic research to prophylactic and therapeutic clinical trials ”. Carmen Gómez explains "this review is focused in the Ankara modified version of vaccinia virus, collects our main contributions from the basic knowledge of the MVA vector biology, both in vitro and in vivo, to its evaluation as a vaccine candidate against HIV/AIDS in clinical trials. The work details the generation and characterization of the recombinant vector called MVA-B and the preclinical data that supported the evaluation of MVA-B as the first prophylactic and therapeutic vaccine against HIV-1 tested in humans in Spain. In addition, they assess the results of clinical trials and future lines of research on which to work considering the latest scientific advances in the field of vaccines against HIV.
In addition, professor Mariano Esteban has been distinguished with the Gold version of the Carracido Medal for his scientific and academic career, the highest award granted by the RANF. The award ceremony of the Carracido medal took place on Thursday, January 16th. In the act, he read his speech focused in "Vaccines as a disease control system: Where we are and where we are going". Afterwards, a painting by the Cantabrian Ricardo Sanjuán has been discovered to emphasize the work of Mariano Esteban as former President of the RANF (2013-2019)
Descubre en el campus de Cantoblanco a científicas premiadas y olvidadas por el Nobel
Entre el 3 y el 14 de febrero se realizará un escape road en las facultades de ciencias y biología donde se presentarán perfiles de mujeres galardonadas con el Nobel en un recorrido en el que los visitantes responderán preguntas sobre las investigadoras.
- Los participantes buscarán, a través de una aplicación móvil, carteles ocultos de grandes científicas que no ganaron el Nobel.
- Este “escape road” es una actividad conjunta organizada por los centros del CSIC del campus de Cantoblanco: (Instituto de Ciencia de Materiales de Madrid, Instituto de Cerámica y Vidrio, Instituto de Catálisis y Petroquímica, Centro Nacional de Biotecnología, Centro de Biología Molecular, Instituto de Ciencias Matemáticas, Instituto de Física Teórica e Instituto de Investigación en Ciencias de la Alimentación.)
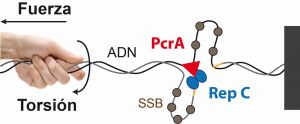
Los plásmidos son moléculas de ADN circulares presentes en bacterias que se replican (duplican) de forma independiente del cromosoma bacteriano. La transmisión de plásmidos entre bacterias puede suponer la ganancia o la perdida de una función determinada en la célula receptora. Por ejemplo, algunos genes contenidos en plásmidos son capaces de conferir resistencia a antibióticos, y la transmisión de estos genes puede ser beneficiosa para la supervivencia bacteriana, ya que se convertirán en resistentes a esos antibióticos.
El sistema más habitual de duplicación de estos plásmidos se llama replicación por circulo rodante (RCR). Este proceso requiere la unión de la proteína Rep a una secuencia determinada del ADN, el corte de una de las cadenas del ADN y la separación de la doble cadena a medida que la helicasa PcrA se mueve por una de ellas. Sin embargo, el mecanismo por el cual se inicia la replicación y se desenrolla la doble hélice del ADN no se conoce con detalle.
Investigadores del Centro Nacional de Biotecnología, en colaboración con un grupo de científicos de la Universidad de Pittsburgh (Estados Unidos) han sido capaces de observar a nivel molecular la interacción de las proteínas Rep y PcrA en el proceso de replicación de plásmidos portadores de genes de resistencia a antibióticos.
COOKIES POLICY
A cookie is a text file that is stored on your computer or mobile device via a web server and only that server will be able to retrieve or read the contents of the cookie and allow the Web site remember browser preferences and navigate efficiently. Cookies make the interaction between the user and the website faster and easier.
General information
This Website uses cookies. Cookies are small text files generated by the web pages you visit, which contain the session data that can be useful later in the website. In this way this Web remembers information about your visit, which can facilitate your next visit and make the website more useful.
How do cookies?
Cookies can only store text, usually always anonymous and encrypted. No personal information is ever stored in a cookie, or can be associated with identified or identifiable person.
The data allow this website to keep your information between the pages, and also to discuss how to interact with the website. Cookies are safe because they can only store information that is put there by the browser, which is information the user entered in the browser or included in the page request. You can not run the code and can not be used to access your computer. If a website encrypts cookie data, only the website can read the information.
What types of cookies used?
The cookies used by this website can be distinguished by the following criteria:
1. Types of cookies as the entity that manages:
Depending on who the entity operating the computer or domain where cookies are sent and treat the data obtained, we can distinguish:
- Own cookies: are those that are sent to the user's terminal equipment from a computer or domain managed by the editor itself and from which provides the service requested by the user.
- Third party cookies: these are those that are sent to the user's terminal equipment from a machine or domain that is not managed by the publisher, but by another entity data is obtained through cookies.
In the event that the cookies are installed from a computer or domain managed by the editor itself but the information collected by these is managed by a third party can not be considered as party cookies.
2. Types of cookies as the length of time that remain active:
Depending on the length of time that remain active in the terminal equipment can be distinguished:
- Session cookies: cookies are a type designed to collect and store data while the user accesses a web page. Are usually used to store information that only worth preserving for the service requested by the user at any one time (eg a list of products purchased).
- Persistent cookies: cookies are a type of data which are stored in the terminal and can be accessed and treated for a period defined by the head of the cookie, and can range from a few minutes to several years.
3. Cookies types according to their purpose:
Depending on the purpose for which the data are processed through cookies, we can distinguish between:
- Technical cookies: these are those that allow the user to navigate through a web page or application platform and the use of different options or services it exist as, for example, control traffic and data communication, identify the session, access to restricted access parts, remember the elements of an order, make the buying process an order, make an application for registration or participation in an event, use security features while browsing store content for dissemination videos or sound or share content via social networks.
- Customization cookies: these are those that allow the user to access the service with some general characteristics based on a predefined set of criteria in the user terminal would eg language, the type of browser through which you access the service, the locale from which you access the service, etc.
- Analysis cookies: they are those that allow the responsible for them, monitoring and analyzing the behavior of users of the web sites that are linked. The information gathered through such cookies are used in measuring the activity of web sites, application or platform and for the profiling of user navigation of such sites, applications and platforms, in order to make improvements function data analysis how users use the service.
Management tool cookies
This Website uses Google Analytics.
Google Analytics is a free tool from Google that primarily allows website owners know how users interact with your website. Also, enable cookies in the domain of the site in which you are and uses a set of cookies called "__utma" and "__utmz" to collect information anonymously and reporting of website trends without identifying individual users..
For statistics of use of this website use cookies in order to know the level of recurrence of our visitors and more interesting content. This way we can concentrate our efforts on improving the most visited areas and make the user more easily find what they are looking for. On this site you can use the information from your visit for statistical evaluations and calculations anonymous data and to ensure the continuity of service or to make improvements to their websites. For more details, see the link below privacy policy [http://www.google.com/intl/en/policies/privacy/]
How to manage cookies on your computer: disabling and deleting cookies
All Internet browsers allow you to limit the behavior of a cookie or disable cookies within settings or browser settings. The steps for doing so are different for each browser, you can find instructions in the help menu of your browser.
If you decline the use of cookies, since it is possible thanks to the preferences menu of your browser or settings, reject, this website will continue to function properly without the use of the same.
Can you allow, block or delete cookies installed on your computer by setting your browser options installed on your computer:
- For more information about Internet Explorer click here.
- For more information on Chrome click here.
- For more information about Safari click here.
- For more information about Firefox click here.
Through your browser, you can also view the cookies that are on your computer, and delete them as you see fit. Cookies are text files, you can open and read the contents. The data within them is almost always encrypted with a numeric key corresponding to an Internet session so often has no meaning beyond the website who wrote it.
Informed consent
The use of this website on the other hand, implies that you paid your specific consent to the use of cookies, on the terms and conditions provided in this Cookies Policy, without prejudice to the measures of deactivation and removal of cookies that you can take, and mentioned in the previous section.