Jaime Martín Benito
Jaime Martín Benito
ULTRASTRUCTURE OF VIRUS AND MOLECULAR AGGREGATES
The main research line of our group is the study of the influenza A ribonucleoproteins (RNPs) that conform the virus nucleocapsid.
Los plásmidos son moléculas de ADN circulares presentes en bacterias que se replican (duplican) de forma independiente del cromosoma bacteriano. La transmisión de plásmidos entre bacterias puede suponer la ganancia o la perdida de una función determinada en la célula receptora. Por ejemplo, algunos genes contenidos en plásmidos son capaces de conferir resistencia a antibióticos, y la transmisión de estos genes puede ser beneficiosa para la supervivencia bacteriana, ya que se convertirán en resistentes a esos antibióticos.
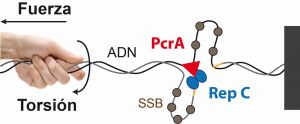
El sistema más habitual de duplicación de estos plásmidos se llama replicación por circulo rodante (RCR). Este proceso requiere la unión de la proteína Rep a una secuencia determinada del ADN, el corte de una de las cadenas del ADN y la separación de la doble cadena a medida que la helicasa PcrA se mueve por una de ellas. Sin embargo, el mecanismo por el cual se inicia la replicación y se desenrolla la doble hélice del ADN no se conoce con detalle.
Investigadores del Centro Nacional de Biotecnología, en colaboración con un grupo de científicos de la Universidad de Pittsburgh (Estados Unidos) han sido capaces de observar a nivel molecular la interacción de las proteínas Rep y PcrA en el proceso de replicación de plásmidos portadores de genes de resistencia a antibióticos.
Nucleic Acids Res. 2020 Jan 13. pii: gkz1200.
Carrasco C, Pastrana CL, Aicart-Ramos C, Leuba SH, Khan SA, Moreno-Herrero F.
Abstract
The rolling-circle replication is the most common mechanism for the replication of small plasmids carrying antibiotic resistance genes in Gram-positive bacteria. It is initiated by the binding and nicking of double-stranded origin of replication by a replication initiator protein (Rep). Duplex unwinding is then performed by the PcrA helicase, whose processivity is critically promoted by its interaction with Rep. How Rep and PcrA proteins interact to nick and unwind the duplex is not fully understood. Here, we have used magnetic tweezers to monitor PcrA helicase unwinding and its relationship with the nicking activity of Staphylococcus aureus plasmid pT181 initiator RepC. Our results indicate that PcrA is a highly processive helicase prone to stochastic pausing, resulting in average translocation rates of 30 bp s-1, while a typical velocity of 50 bp s-1 is found in the absence of pausing. Single-strand DNA binding protein did not affect PcrA translocation velocity but slightly increased its processivity. Analysis of the degree of DNA supercoiling required for RepC nicking, and the time between RepC nicking and DNA unwinding, suggests that RepC and PcrA form a protein complex on the DNA binding site before nicking. A comprehensive model that rationalizes these findings is presented.
doi: 10.1093/nar/gkz1200.
Nat Commun. 2020 Jan 2;11(1):55.
Vilas JL, Tagare HD, Vargas J, Carazo JM, Sorzano COS.
Abstract
The introduction of local resolution has enormously helped the understanding of cryo-EM maps. Still, for any given pixel it is a global, aggregated value, that makes impossible the individual analysis of the contribution of the different projection directions. We introduce MonoDir, a fully automatic, parameter-free method that, starting only from the final cryo-EM map, decomposes local resolution into the different projection directions, providing a detailed level of analysis of the final map. Many applications of directional local resolution are possible, and we concentrate here on map quality and validation.
doi: 10.1038/s41467-019-13742-w.
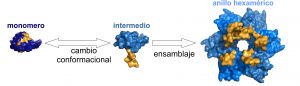
Muchas macromoléculas biológicas utilizan sistemas de control alostérico para ensamblarse, desensamblarse o cambiar de conformación. Así, cambios en una región de la molécula afectan al comportamiento y la función de la misma en otras regiones. Por ejemplo, la unión de un ligando produce un cambio conformacional que activa o inactiva una enzima.
El desarrollo de moléculas sintéticas que tengan este tipo de propiedades resulta muy complejo y hasta ahora los esfuerzos invertidos en este área solo han conseguido unos pocos resultados exitosos. De hecho, en el caso concreto de la creación de nuevos ensamblajes protéicos de diseño, los resultados hasta ahora se han basado en un sistema tipo puzzle, donde la superficie de cada una de las piezas rígidas que se van a ensamblar es modificada para producir una interacción estable. Ahora, investigadores del Centro Nacional de Biotecnología, en colaboración con el IMDEA Nanociencia, el Centro de Investigaciones Biológicas y las Universidades de Granada y California han desarrollado un modelo que permite controlar de forma alostérica la formación y ensamblaje de proteínas mediante la inclusión de interruptores moleculares formados gracias a cambios en la estructura tridimensional de la proteína.
Sachse M, Fernández de Castro I, Tenorio R, Risco C.
Abstract
Transmission electron microscopy (TEM) has been crucial to study viral infections. As a result of recent advances in light and electron microscopy, we are starting to be aware of the variety of structures that viruses assemble inside cells. Viruses often remodel cellular compartments to build their replication factories. Remarkably, viruses are also able to induce new membranes and new organelles. Here we revise the most relevant imaging technologies to study the biogenesis of viral replication organelles. Live cell microscopy, correlative light and electron microscopy, cryo-TEM, and three-dimensional imaging methods are unveiling how viruses manipulate cell organization. In particular, methods for molecular mapping in situ in two and three dimensions are revealing how macromolecular complexes build functional replication complexes inside infected cells. The combination of all these imaging approaches is uncovering the viral life cycle events with a detail never seen before.
doi: 10.1016/bs.aivir.2019.07.005.
Nucleic Acids Res. 2019 Aug 9.
Martín-González N, Hernando-Pérez M, Condezo GN, Pérez-Illana M, Šiber A, Reguera D, Ostapchuk P, Hearing P6, San Martín C, de Pablo PJ.
Abstract
Some viruses package dsDNA together with large amounts of positively charged proteins, thought to help condense the genome inside the capsid with no evidence. Further, this role is not clear because these viruses have typically lower packing fractions than viruses encapsidating naked dsDNA. In addition, it has recently been shown that the major adenovirus condensing protein (polypeptide VII) is dispensable for genome encapsidation. Here, we study the morphology and mechanics of adenovirus particles with (Ad5-wt) and without (Ad5-VII-) protein VII. Ad5-VII- particles are stiffer than Ad5-wt, but DNA-counterions revert this difference, indicating that VII screens repulsive DNA-DNA interactions. Consequently, its absence results in increased internal pressure. The core is slightly more ordered in the absence of VII and diffuses faster out of Ad5-VII- than Ad5-wt fractured particles. In Ad5-wt unpacked cores, dsDNA associates in bundles interspersed with VII-DNA clusters. These results indicate that protein VII condenses the adenovirus genome by combining direct clustering and promotion of bridging by other core proteins. This condensation modulates the virion internal pressure and DNA release from disrupted particles, which could be crucial to keep the genome protected inside the semi-disrupted capsid while traveling to the nuclear pore.
doi: 10.1093/nar/gkz687
COOKIES POLICY
A cookie is a text file that is stored on your computer or mobile device via a web server and only that server will be able to retrieve or read the contents of the cookie and allow the Web site remember browser preferences and navigate efficiently. Cookies make the interaction between the user and the website faster and easier.
General information
This Website uses cookies. Cookies are small text files generated by the web pages you visit, which contain the session data that can be useful later in the website. In this way this Web remembers information about your visit, which can facilitate your next visit and make the website more useful.
How do cookies?
Cookies can only store text, usually always anonymous and encrypted. No personal information is ever stored in a cookie, or can be associated with identified or identifiable person.
The data allow this website to keep your information between the pages, and also to discuss how to interact with the website. Cookies are safe because they can only store information that is put there by the browser, which is information the user entered in the browser or included in the page request. You can not run the code and can not be used to access your computer. If a website encrypts cookie data, only the website can read the information.
What types of cookies used?
The cookies used by this website can be distinguished by the following criteria:
1. Types of cookies as the entity that manages:
Depending on who the entity operating the computer or domain where cookies are sent and treat the data obtained, we can distinguish:
- Own cookies: are those that are sent to the user's terminal equipment from a computer or domain managed by the editor itself and from which provides the service requested by the user.
- Third party cookies: these are those that are sent to the user's terminal equipment from a machine or domain that is not managed by the publisher, but by another entity data is obtained through cookies.
In the event that the cookies are installed from a computer or domain managed by the editor itself but the information collected by these is managed by a third party can not be considered as party cookies.
2. Types of cookies as the length of time that remain active:
Depending on the length of time that remain active in the terminal equipment can be distinguished:
- Session cookies: cookies are a type designed to collect and store data while the user accesses a web page. Are usually used to store information that only worth preserving for the service requested by the user at any one time (eg a list of products purchased).
- Persistent cookies: cookies are a type of data which are stored in the terminal and can be accessed and treated for a period defined by the head of the cookie, and can range from a few minutes to several years.
3. Cookies types according to their purpose:
Depending on the purpose for which the data are processed through cookies, we can distinguish between:
- Technical cookies: these are those that allow the user to navigate through a web page or application platform and the use of different options or services it exist as, for example, control traffic and data communication, identify the session, access to restricted access parts, remember the elements of an order, make the buying process an order, make an application for registration or participation in an event, use security features while browsing store content for dissemination videos or sound or share content via social networks.
- Customization cookies: these are those that allow the user to access the service with some general characteristics based on a predefined set of criteria in the user terminal would eg language, the type of browser through which you access the service, the locale from which you access the service, etc.
- Analysis cookies: they are those that allow the responsible for them, monitoring and analyzing the behavior of users of the web sites that are linked. The information gathered through such cookies are used in measuring the activity of web sites, application or platform and for the profiling of user navigation of such sites, applications and platforms, in order to make improvements function data analysis how users use the service.
Management tool cookies
This Website uses Google Analytics.
Google Analytics is a free tool from Google that primarily allows website owners know how users interact with your website. Also, enable cookies in the domain of the site in which you are and uses a set of cookies called "__utma" and "__utmz" to collect information anonymously and reporting of website trends without identifying individual users..
For statistics of use of this website use cookies in order to know the level of recurrence of our visitors and more interesting content. This way we can concentrate our efforts on improving the most visited areas and make the user more easily find what they are looking for. On this site you can use the information from your visit for statistical evaluations and calculations anonymous data and to ensure the continuity of service or to make improvements to their websites. For more details, see the link below privacy policy [http://www.google.com/intl/en/policies/privacy/]
How to manage cookies on your computer: disabling and deleting cookies
All Internet browsers allow you to limit the behavior of a cookie or disable cookies within settings or browser settings. The steps for doing so are different for each browser, you can find instructions in the help menu of your browser.
If you decline the use of cookies, since it is possible thanks to the preferences menu of your browser or settings, reject, this website will continue to function properly without the use of the same.
Can you allow, block or delete cookies installed on your computer by setting your browser options installed on your computer:
- For more information about Internet Explorer click here.
- For more information on Chrome click here.
- For more information about Safari click here.
- For more information about Firefox click here.
Through your browser, you can also view the cookies that are on your computer, and delete them as you see fit. Cookies are text files, you can open and read the contents. The data within them is almost always encrypted with a numeric key corresponding to an Internet session so often has no meaning beyond the website who wrote it.
Informed consent
The use of this website on the other hand, implies that you paid your specific consent to the use of cookies, on the terms and conditions provided in this Cookies Policy, without prejudice to the measures of deactivation and removal of cookies that you can take, and mentioned in the previous section.