Plant Cell Environ. 2024 Mar 18.
López B, Izquierdo Y, Cascón T, Zamarreño ÁM, García-Mina JM, Pulido P, Castresana C.
Abstract
Chloroplast function is essential for growth, development, and plant adaptation to stress. Organelle stress and plant defence responses were examined here using noxy8 (nonresponding to oxylipins 8) from a series of Arabidopsis mutants. The noxy8 mutation was located at the CLPC2 gene, encoding a chloroplast chaperone of the protease complex CLP. Although its CLPC1 paralogue is considered to generate redundancy, our data reveal significant differences distinguishing CLPC2 and CLPC1 functions. As such, clpc1 mutants displayed a major defect in housekeeping chloroplast proteostasis, leading to a pronounced reduction in growth and pigment levels, enhanced accumulation of chloroplast and cytosol chaperones, and resistance to fosmidomycin. Conversely, clpc2 mutants showed severe susceptibility to lincomycin inhibition of chloroplast translation and resistance to Antimycin A inhibition of mitochondrial respiration. In the response to Pseudomonas syringae pv. tomato, clpc2 but not clpc1 mutants were resistant to bacterial infection, showing higher salicylic acid levels, defence gene expression and 9-LOX pathway activation. Our findings suggest CLPC2 and CLPC1 functional specificity, with a preferential involvement of CLPC1 in housekeeping processes and of CLPC2 in stress responses.
DOI: 10.1111/pce.14882
Plant Cell Environ. 2022 Mar 15.
Noelia Arteaga, Belén Méndez-Vigo, Alberto Fuster-Pons, Marija Savic, Alba Murillo-Sánchez, F Xavier Picó, Carlos Alonso-Blanco
Abstract
Despite the adaptive and taxonomic relevance of the natural diversity for trichome patterning and morphology, the molecular and evolutionary mechanisms underlying these traits remain mostly unknown, particularly in organs other than leaves. In this study, we address the ecological, genetic and molecular bases of the natural variation for trichome patterning and branching in multiple organs of Arabidopsis (Arabidopsis thaliana). To this end, we characterized a collection of 191 accessions and carried out environmental and genome-wide association (GWA) analyses. Trichome amount in different organs correlated negatively with precipitation in distinct seasons, thus suggesting a precise fit between trichome patterning and climate throughout the Arabidopsis life cycle. In addition, GWA analyses showed small overlapping between the genes associated with different organs, indicating partly independent genetic bases for vegetative and reproductive phases. These analyses identified a complex locus on chromosome 2, where two adjacent MYB genes (ETC2 and TCL1) displayed differential effects on trichome patterning in several organs. Furthermore, analyses of transgenic lines carrying different natural alleles demonstrated that TCL1 accounts for the variation for trichome patterning in all organs, and for stem trichome branching. By contrast, two other MYB genes (TRY and GL1), mainly showed effects on trichome patterning or branching, respectively.
doi: 10.1111/pce.14308. Online ahead of print.
The Plant Cell, 4 Jan 2021, koaa041
Noelia Arteaga, Marija Savic, Belén Méndez-Vigo, Alberto Fuster-Pons, Rafael Torres-Pérez, Juan Carlos Oliveros, F Xavier Picó, Carlos Alonso-Blanco
Abstract
Both inter- and intra-specific diversity has been described for trichome patterning in fruits, which is presumably involved in plant adaptation. However, the mechanisms underlying this developmental trait have been hardly addressed. Here we examined natural populations of Arabidopsis (Arabidopsis thaliana) that develop trichomes in fruits and pedicels, phenotypes previously not reported in the Arabidopsis genus. Genetic analyses identified five loci, MALAMBRUNO 1–5 (MAU1–5), with MAU2, MAU3, and MAU5 showing strong epistatic interactions that are necessary and sufficient to display these traits. Functional characterization of these three loci revealed cis-regulatory mutations in TRICHOMELESS1 and TRIPTYCHON, as well as a structural mutation in GLABRA1. Therefore, the multiple mechanisms controlled by three MYB transcription factors of the core regulatory network for trichome patterning have jointly been modulated to trigger trichome development in fruits. Furthermore, analyses of worldwide accessions showed that these traits and mutations only occur in a highly differentiated relict lineage from the Iberian Peninsula. In addition, these traits and alleles were associated with low spring precipitation, which suggests that trichome development in fruits and pedicels might be involved in climatic adaptation. Thus, we show that the combination of synergistic mutations in a gene regulatory circuit has driven evolutionary innovations in fruit trichome patterning in Arabidopsis.
The EMBO Journal (2018) e99552
Cristina Martínez, Ana Espinosa‐Ruíz, Miguel de Lucas, Stella Bernardo‐García, José M Franco‐Zorrilla, Salomé Prat.
Abstract
The Arabidopsis PIF4 and BES1/BZR1 transcription factors antagonize light signaling by facilitating co‐activated expression of a large number of cell wall‐loosening and auxin‐related genes. While PIF4 directly activates expression of these targets, BES1 and BZR1 activity switch from a repressive to an activator function, depending on interaction with TOPLESS and other families of regulators including PIFs. However, the complexity of this regulation and its role in diurnal control of plant growth and brassinosteroid (BR) levels is little understood. We show by using a protein array that BES1, PIF4, and the BES1‐PIF4 complex recognize different DNA elements, thus revealing a distinctive cis‐regulatory code beneath BES1‐repressive and PIF4 co‐activation function. BES1 homodimers bind to conserved BRRE‐ and G‐box elements in the BR biosynthetic promoters and inhibit their expression during the day, while elevated PIF4 competes for BES1 homodimer formation, resulting in de‐repressed BR biosynthesis at dawn and in response to warmth. Our findings demonstrate a central role of PIF4 in BR synthesis activation, increased BR levels being essential to thermomorphogenic hypocotyl growth.
Nature Plants 2015; doi: 10.1038/nplants.2015.25.
Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, Miñambres M, Walther D, Schulze WX, Paz-Ares J, Scheible WR, Kragler F.
 The concept that proteins and small RNAs can move to and function in distant body parts is well established. However, non-cell-autonomy of small RNA molecules raises the question: To what extent are protein-coding messenger RNAs (mRNAs) exchanged between tissues in plants?
The concept that proteins and small RNAs can move to and function in distant body parts is well established. However, non-cell-autonomy of small RNA molecules raises the question: To what extent are protein-coding messenger RNAs (mRNAs) exchanged between tissues in plants?
Here we report the comprehensive identification of 2,006 genes producing mobile RNAs in Arabidopsis thaliana. The analysis of variant ecotype transcripts that were present in heterografted plants allowed the identification of mRNAs moving between various organs under normal or nutrient-limiting conditions. Most of these mobile transcripts seem to follow the phloem-dependent allocation pathway transporting sugars from photosynthetic tissues to roots via the vasculature. Notably, a high number of transcripts also move in the opposite, root-to-shoot direction and are transported to specific tissues including flowers. Proteomic data on grafted plants indicate the presence of proteins from mobile RNAs, allowing the possibility that they may be translated at their destination site. The mobility of a high number of mRNAs suggests that a postulated tissue-specific gene expression profile might not be predictive for the actual plant body part in which a transcript exerts its function.
Bio-protocol. 2014; 4 (20): e1261.
Chevalier F, Iglesias S M, Sánchez ÓJ, Montoliu L, Cubas P.
 The inflorescence stem of the flowering plant Arabidopsis thaliana (thale cress) is an excellent model system to investigate plant vascular tissue patterning and development. Plant vasculature is a complex conducting tissue arranged in strands called vascular bundles, formed by xylem (tissue that carries water) and phloem (tissue that carries photosynthates and signaling molecules). Xylem and phloem are originated from cell division of the meristematic cells of the vascular cambium. In Arabidopsis the flowering stem elongates about three weeks after germination. At this stage it is possible to visualize defects in its development and morphology.
The inflorescence stem of the flowering plant Arabidopsis thaliana (thale cress) is an excellent model system to investigate plant vascular tissue patterning and development. Plant vasculature is a complex conducting tissue arranged in strands called vascular bundles, formed by xylem (tissue that carries water) and phloem (tissue that carries photosynthates and signaling molecules). Xylem and phloem are originated from cell division of the meristematic cells of the vascular cambium. In Arabidopsis the flowering stem elongates about three weeks after germination. At this stage it is possible to visualize defects in its development and morphology.
Here we describe a protocol to embed in plastic (resin) stem segments either freshly dissected from living plants or previously assayed for β-glucuronidase. This protocol provides an excellent cellular morphology ideal to visualize stem cell types including those of vascular bundles using high-resolution light microscopy.
Plant Cell. 2014, pii: tpc.114.125047.
Chico JM, Fernández-Barbero G, Chini A, Fernández-Calvo P, Díez-Díaz M, Solano R.
 Reduction of the red/far-red (R/FR) light ratio that occurs in dense canopies promotes plant growth to outcompete neighbors but has a repressive effect on jasmonate (JA)–dependent defenses. The molecular mechanism underlying this trade-off is not well understood.
Reduction of the red/far-red (R/FR) light ratio that occurs in dense canopies promotes plant growth to outcompete neighbors but has a repressive effect on jasmonate (JA)–dependent defenses. The molecular mechanism underlying this trade-off is not well understood.
We found that the JA-related transcription factors MYC2, MYC3, and MYC4 are short-lived proteins degraded by the proteasome, and stabilized by JA and light, in Arabidopsis thaliana. Dark and CONSTITUTIVE PHOTOMORPHOGENIC1 destabilize MYC2, MYC3, and MYC4, whereas R and blue (B) lights stabilize them through the activation of the corresponding photoreceptors. Consistently, phytochrome B inactivation by monochromatic FR light or shade (FR-enriched light) destabilizes these three proteins and reduces their stabilization by JA. In contrast to MYCs, simulated shade conditions stabilize seven of their 10 JAZ repressors tested and reduce their degradation by JA.
MYC2, MYC3, and MYC4 are required for JA-mediated defenses against the necrotrophic pathogen Botrytis cinerea and for the shade-triggered increased susceptibility, indicating that this negative effect of shade on defense is likely mediated by shade-triggered inactivation of MYC2, MYC3, and MYC4. The opposite regulation of protein stability of MYCs and JAZs by FR-enriched light help explain (on the molecular level) the long-standing observation that canopy shade represses JA-mediated defenses, facilitating reallocation of resources from defense to growth.
Las bacterias que atacan a las plantas, además de secretar toxinas, inyectan en las células vegetales proteínas efectoras que favorecen la infección. Algunas cepas de Pseudomonas producen una toxina llamada coronatina que desorganiza la fisiología de la célula e impide el funcionamiento de sus defensas. La coronatina imita el funcionamiento de una de las hormonas encargadas de activar las defensas contra los hongos, el jasmonato. Pero, al mismo tiempo, promueve el crecimiento bacteriano al inhibir los mecanismos de defensa dependientes del ácido salicílico, los cuales son fundamentales para resitir las infecciones por Pseudomonas.
El grupo dirigido por Roberto Solano en el Centro Nacional de Biotecnología del CSIC intenta elucidar cómo las cepas de Pseudomonas que no producen coronatina, la mayoría de ellas, son capaces de alterar las hormonas de las plantas y qué mecanismos han evolucionado para que puedan infectarlas.
Aparte de las fitotoxinas, las cepas de Pseudomonas producen hasta 30 proteínas efectoras de tipo III distintas que son capaces de suprimir de forma activa las defensas de las plantas infectadas. Para el caso concreto de uno de estos efectores, HopX1, Solano acaba de encontar en su laboratorio del CNB que en la planta modelo Arabidopsis, tanto la expresión exógena de HopX1 como la infección natural por cepas de Pseudomonas que producen este efector, promueven el crecimeinto bacteriano de manera similar a la coronatina: activando la expresión de los genes dependientes del jasmonato. Sus resultados indican que HopX1 es una cisteína proteasa que imita los efectos de la coronatina al degradar a represores esenciales en el funcionamiento de la vía de señalización del jasmonato, las llamadas proteínas jasmonate-ZIM domain (JAZ).
Estos resultados, publicados en la revista PLoS Biology, muestran un mecanismo hasta ahora desconocido por el que los patógenos manipulan los puntos clave de la respuesta hormonal que usan las plantas para protegerse frente a las infecciones.
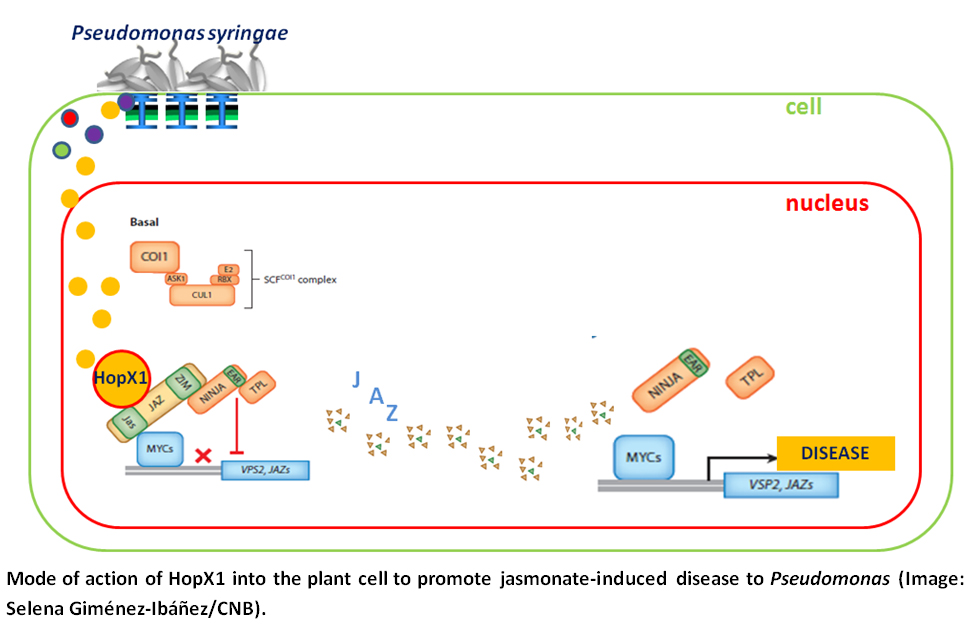
- Gimenez-Ibanez S, Boter M, Fernández-Barbero G, Chini A, Rathjen JP, Solano R. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 2014; 12(2): e1001792.
Plant Cell. 2014; 26 (2) :712-728.
Irigoyen ML1, Iniesto E, Rodriguez L, Puga MI, Yanagawa Y, Pick E, Strickland E, Paz-Ares J, Wei N, De Jaeger G, Rodriguez PL, Deng XW, Rubio V.
 CULLIN4-RING E3 ubiquitin ligases (CRL4s) regulate key developmental and stress responses in eukaryotes. Studies in both animals and plants have led to the identification of many CRL4 targets as well as specific regulatory mechanisms that modulate their function. The latter involve COP10-DET1-DDB1 (CDD)–related complexes, which have been proposed to facilitate target recognition by CRL4, although the molecular basis for this activity remains largely unknown.
CULLIN4-RING E3 ubiquitin ligases (CRL4s) regulate key developmental and stress responses in eukaryotes. Studies in both animals and plants have led to the identification of many CRL4 targets as well as specific regulatory mechanisms that modulate their function. The latter involve COP10-DET1-DDB1 (CDD)–related complexes, which have been proposed to facilitate target recognition by CRL4, although the molecular basis for this activity remains largely unknown.
Here, we provide evidence that Arabidopsis thaliana DET1-, DDB1-ASSOCIATED1 (DDA1), as part of the CDD complex, provides substrate specificity for CRL4 by interacting with ubiquitination targets. Thus, we show that DDA1 binds to the abscisic acid (ABA) receptor PYL8, as well as PYL4 and PYL9, in vivo and facilitates its proteasomal degradation. Accordingly, we found that DDA1 negatively regulates ABA-mediated developmental responses, including inhibition of seed germination, seedling establishment, and root growth. All other CDD components displayed a similar regulatory function, although they did not directly interact with PYL8. Interestingly, DDA1-mediated destabilization of PYL8 is counteracted by ABA, which protects PYL8 by limiting its polyubiquitination.
Altogether, our data establish a function for DDA1 as a substrate receptor for CRL4-CDD complexes and uncover a mechanism for the desensitization of ABA signaling based on the regulation of ABA receptor stability.
COOKIES POLICY
A cookie is a text file that is stored on your computer or mobile device via a web server and only that server will be able to retrieve or read the contents of the cookie and allow the Web site remember browser preferences and navigate efficiently. Cookies make the interaction between the user and the website faster and easier.
General information
This Website uses cookies. Cookies are small text files generated by the web pages you visit, which contain the session data that can be useful later in the website. In this way this Web remembers information about your visit, which can facilitate your next visit and make the website more useful.
How do cookies?
Cookies can only store text, usually always anonymous and encrypted. No personal information is ever stored in a cookie, or can be associated with identified or identifiable person.
The data allow this website to keep your information between the pages, and also to discuss how to interact with the website. Cookies are safe because they can only store information that is put there by the browser, which is information the user entered in the browser or included in the page request. You can not run the code and can not be used to access your computer. If a website encrypts cookie data, only the website can read the information.
What types of cookies used?
The cookies used by this website can be distinguished by the following criteria:
1. Types of cookies as the entity that manages:
Depending on who the entity operating the computer or domain where cookies are sent and treat the data obtained, we can distinguish:
- Own cookies: are those that are sent to the user's terminal equipment from a computer or domain managed by the editor itself and from which provides the service requested by the user.
- Third party cookies: these are those that are sent to the user's terminal equipment from a machine or domain that is not managed by the publisher, but by another entity data is obtained through cookies.
In the event that the cookies are installed from a computer or domain managed by the editor itself but the information collected by these is managed by a third party can not be considered as party cookies.
2. Types of cookies as the length of time that remain active:
Depending on the length of time that remain active in the terminal equipment can be distinguished:
- Session cookies: cookies are a type designed to collect and store data while the user accesses a web page. Are usually used to store information that only worth preserving for the service requested by the user at any one time (eg a list of products purchased).
- Persistent cookies: cookies are a type of data which are stored in the terminal and can be accessed and treated for a period defined by the head of the cookie, and can range from a few minutes to several years.
3. Cookies types according to their purpose:
Depending on the purpose for which the data are processed through cookies, we can distinguish between:
- Technical cookies: these are those that allow the user to navigate through a web page or application platform and the use of different options or services it exist as, for example, control traffic and data communication, identify the session, access to restricted access parts, remember the elements of an order, make the buying process an order, make an application for registration or participation in an event, use security features while browsing store content for dissemination videos or sound or share content via social networks.
- Customization cookies: these are those that allow the user to access the service with some general characteristics based on a predefined set of criteria in the user terminal would eg language, the type of browser through which you access the service, the locale from which you access the service, etc.
- Analysis cookies: they are those that allow the responsible for them, monitoring and analyzing the behavior of users of the web sites that are linked. The information gathered through such cookies are used in measuring the activity of web sites, application or platform and for the profiling of user navigation of such sites, applications and platforms, in order to make improvements function data analysis how users use the service.
Management tool cookies
This Website uses Google Analytics.
Google Analytics is a free tool from Google that primarily allows website owners know how users interact with your website. Also, enable cookies in the domain of the site in which you are and uses a set of cookies called "__utma" and "__utmz" to collect information anonymously and reporting of website trends without identifying individual users..
For statistics of use of this website use cookies in order to know the level of recurrence of our visitors and more interesting content. This way we can concentrate our efforts on improving the most visited areas and make the user more easily find what they are looking for. On this site you can use the information from your visit for statistical evaluations and calculations anonymous data and to ensure the continuity of service or to make improvements to their websites. For more details, see the link below privacy policy [http://www.google.com/intl/en/policies/privacy/]
How to manage cookies on your computer: disabling and deleting cookies
All Internet browsers allow you to limit the behavior of a cookie or disable cookies within settings or browser settings. The steps for doing so are different for each browser, you can find instructions in the help menu of your browser.
If you decline the use of cookies, since it is possible thanks to the preferences menu of your browser or settings, reject, this website will continue to function properly without the use of the same.
Can you allow, block or delete cookies installed on your computer by setting your browser options installed on your computer:
- For more information about Internet Explorer click here.
- For more information on Chrome click here.
- For more information about Safari click here.
- For more information about Firefox click here.
Through your browser, you can also view the cookies that are on your computer, and delete them as you see fit. Cookies are text files, you can open and read the contents. The data within them is almost always encrypted with a numeric key corresponding to an Internet session so often has no meaning beyond the website who wrote it.
Informed consent
The use of this website on the other hand, implies that you paid your specific consent to the use of cookies, on the terms and conditions provided in this Cookies Policy, without prejudice to the measures of deactivation and removal of cookies that you can take, and mentioned in the previous section.