Álvaro Sánchez (until September 2023)
Group Leader
Research summary
In the Microbial E&E lab we study the evolution and assembly of microbial consortia, and how their collective functions emerge from a complex web of interactions between their components
Publications
J Diaz-Colunga, N Lu, A Sanchez-Gorostiaga, C-Y Chang, H Cai, J Goldford, M Tikhonov & A Sanchez. Top-down and bottom-up cohesiveness in microbial community coalescence. Proceedings of the National Academy of Sciences USA (2022) 119(6):e2111261119
S Estrela, JC. C. Vila, N Lu, D Bajic, M Rebolleda-Gomez, C-Y Chang & A Sanchez. Metabolic rules of microbial community assembly. Cell Systems (2021)
Chang CY, Vila JCC, Bender M, Li R, Mankowski MC, Bassette M, Borden J, Golfier S, Sanchez S, Waymack R, Zhu X, Diaz-Colunga J, Estrela S, Rebolleda-Gomez M & Sanchez A. Engineering complex communities by directed evolution. Nature Ecology & Evolution (2021) 5, 1011–1023
S Estrela, A Sanchez-Gorostiaga, JC Vila & A Sanchez. Nutrient dominance governs the assembly of microbial communities in mixed nutrient environments. eLife (2021) 10:e65948
A Sanchez-Gorostiaga, D Bajic, ML Osborne, JF Poyatos, Sanchez, A. High-order interactions dominate the functional landscape of microbial consortia. PLoS Biology (2019) 17(12): e3000550
J Goldford, N Lu, D Bajic, S Estrela, A Sanchez-Gorostiaga, M Tikhonov, D Segre, P Mehta, A Sanchez. Emergent simplicity in microbial community assembly. Science (2018) 361:469-74
Our research centers on exploring how evolutionary biology may help us make sense, predict, and engineer microbial communities. To this end, we employ a combination of laboratory experiments, computer simulations, and mathematical modeling. Our laboratory is highly interdisciplinary and quantitative in nature, and our students and postdocs typically combine theory and experiment in their projects, acquiring a broad range of technical skills across the wet-lab / dry-lab divide. They also get exposed to a broad scientific intellectual background in ecology, evolution and systems biology. Our current lines of work are described below.
1) Engineering microbial consortia from the top-down. For millennia, selective breeding has allowed us to artificially select crops and domestic animals. In more recent times, the idea of directed evolution has been extended to engineering biological systems at or below the organismal level: from enzymes and RNA molecules to genetic circuits, metabolic networks, and microbial strains. Can these approaches be also extended to engineering biological systems above the organismal level of organization, such as microbial consortia? Our laboratory is exploring how one may do this successfully. We seek to develop empirical methods, grounded on ecological and evolutionary theory, that may be then adapted by applied biotechnologists in their particular settings. If you wish to know more about this line of work, see our recent publications by Diaz-Colunga (2022), Chang et al (2021), Sanchez et al (2021) and Chang et al (2020)
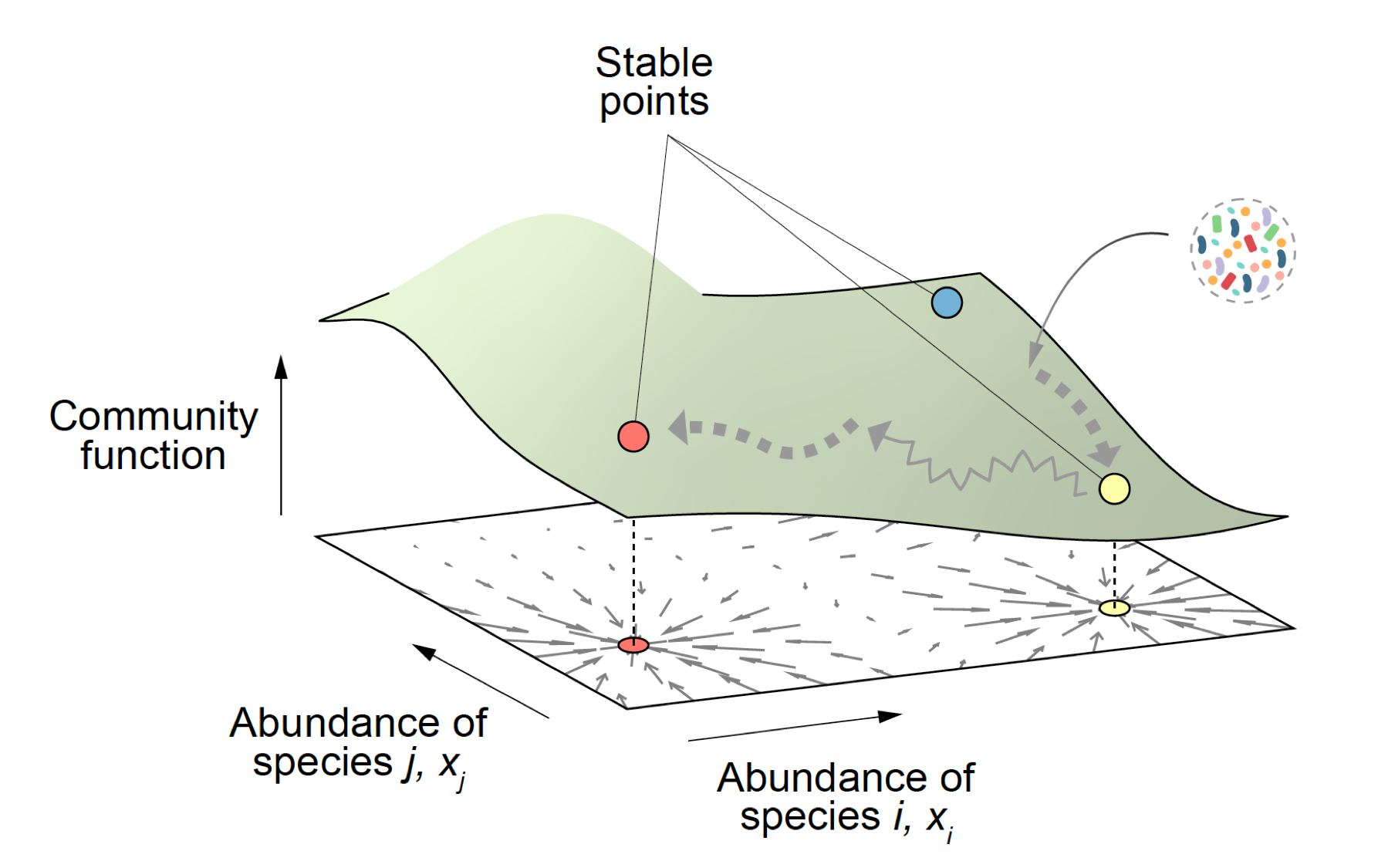
2) Predictively linking microbial community composition and community function. Microbial communities provide us with countless ecological services that are essential for the health of the biosphere and to preserve life as we know it. They also carry out a vast and growing number of important functions in biotechnology, from food production to the synthesis of biofuels and other economically critical molecules. These community functions are determined by community composition, i.e. by which genotypes are found in each community and their abundance. If we wish to engineer microbial consortia to optimize the functions they provide, it is key that we predictively and quantitatively link community composition and function. This is a hard problem, that cannot be solved empirically due to the astronomical number of potential consortia one may form even with a modest number of potential candidate genotypes. Microscopic models are exceedingly difficult to build, due to the complex web of interactions that are involved. Our laboratory is exploring whether ideas from the theory of fitness landscapes in genetics might help us solve this problem and produce predictive and quantitative models of community function from its composition. If you wish to learn more about this line of work, see our recent publications by Sanchez et al (2022), Diaz-Colunga et al (2022), Lino et al (2021), Sanchez (2020), Sanchez-Gorostiaga et al (2019)
3) Microbial evolution and community ecology. Although our main ongoing research lines are the two described above, we maintain a general interest in broad topics in microbial evolution and community ecology. For instance, what factors govern microbial biodiversity? How well can pairwise vs higher-order models predict coexistence and exclusion? Under which conditions will evolution contribute more genetic diversity than ecological processes, such as migration? How does evolution affect the stability of community functions? These are just a sample of questions of current interest. In the past, we have addressed a gamut of questions regarding the reproducibility and predictability of microbial community assembly at the functional and taxonomic scales and the effect of niche construction in microbial evolution, For more information about these completed projects, you may want to read Estrela et al (2022), Estrela et al (2021), Dukovski et al (2021), Vila et al (2020), Marsland et al (2019), Bajic et al (2018) or Goldford et al (2018).
- La adquisición de mutaciones de resistencia a antibióticos puede dar lugar a la sensibilización frente a otros antibióticos, un fenómeno conocido como sensibilidad colateral
- Comprender la evolución de la resistencia a los antibióticos podría facilitar el diseño de nuevas estrategias terapéuticas utilizando antibióticos de uso habitual
La resistencia de las bacterias a los antibióticos es, tal como indican la OMS y la ONU, uno de los mayores problemas actuales de salud pública. Dicha resistencia hace más difícil -en ocasiones imposible- tratar las infecciones bacterianas, así como desarrollar prácticas clínicas como trasplantes o tratamientos anticancerígenos que requieren un control de posibles infecciones. Entre estas, tienen gran relevancia las causadas por Pseudomonas aeruginosa, uno de los patógenos oportunistas que con mayor frecuencia produce infecciones en pacientes hospitalizados o con patologías previas, incluyendo inmunodeficiencias y fibrosis quística. Esta bacteria tiene una baja sensibilidad a los antibióticos, y una gran capacidad para adquirir mutaciones de resistencia a los mismos, de ahí el interés en encontrar terapias eficaces contra ella. En este contexto, además de desarrollar nuevos antibióticos, algo económicamente costoso, es necesario hacer un mejor uso de los que ya tenemos.
Un nuevo estudio de los investigadores del CSIC Sara Hernando-Amado, Pablo Laborda, José Ramón Valverde y José Luis Martínez publicado en la revista Proceedings of the National Academy of Sciences (PNAS), da un paso más en la búsqueda de alternativas terapéuticas basadas en el uso de antibióticos ya existentes. Analizando la evolución de diferentes mutantes de P. aeruginosa en presencia de distintos antibióticos, han identificado un patrón conservado de sensibilidad frente a otros antibióticos (un fenómeno llamado sensibilidad colateral) que podría tener aplicación clínica.
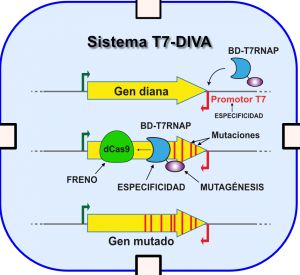
- Las técnicas que permiten acelerar la evolución de proteínas son herramientas muy útiles en procesos de biomedicina y biotecnología
- El nuevo sistema T7-DIVA, diseñado por investigadores del CNB-CSIC, permite el rápido desarrollo de variedades sintéticas de anticuerpos y proteínas relacionadas con la resistencia a antibióticos entre otras aplicaciones
Investigadores del Centro Nacional de Biotecnología del Consejo Superior de Investigaciones Científicas (CNB-CSIC) desarrollan un nuevo sistema de mutagénesis in vivo que permite generar variantes de proteínas de una forma rápida y sencilla en bacterias, y se puede adaptar para su utilización en levaduras y otras células eucariotas. El nuevo sistema, publicado en Nature Communications, permite la selección de variantes de una forma continua y con poca manipulación.
Las técnicas de evolución molecular dirigida en el laboratorio permiten generar variantes de proteínas con interés biotecnológico o biomédico como anticuerpos o enzimas terapéuticas con funciones mejoradas o incluso nuevas de manera mucho más eficaz. Hasta hace poco tiempo, la mayoría de las técnicas de evolución dirigida se realizaban in vitro, en procesos lentos y tediosos, por lo que en los últimos años hay un interés creciente por el desarrollo de técnicas de evolución molecular in vivo.
Ahora, la nueva técnica desarrollada por el grupo del investigador Luis Ángel Fernández en el Centro Nacional de Biotecnología del CSIC permite dirigir las mutaciones de manera específica a la región génica de interés dentro de las células, expresar las variantes y seleccionarlas de una forma rápida, continua y sin mucha manipulación.
 Alvaro San Millán
Alvaro San Millán
In the Plasmid Biology and Evolution lab we study the role of plasmids as catalysts of bacterial evolution, with a special focus on the evolution of plasmid-mediated antibiotic resistance.
 Rafael Giraldo
Rafael Giraldo
We create, through bottom-up Synthetic Biology, bio-resources based on bacterial amyloids with two major aims: i) understanding the molecular determinants of the shift between function and toxicity in natural amyloids; and ii) generating new devices based on amyloids as constructive resources in Biotechnology and Biomedicine.
- The low affinity of PBP3SAL for current beta-lactam antibiotics might explain why antibiotic therapy is not sufficient to eradicate salmonellosis even after symptoms subside.
Up to 10% of patients with salmonellosis infections may have relapses following treatment with third-generation cephalosporins and remission of initial symptoms. Strikingly, the bacterial strains isolated from patients during the relapse maintain their susceptibility to the antibiotic used in the first place. This anomaly suggests that Salmonella enterica might remain in intracellular locations in which drug accessibility is reduced.
Beta-lactam antibiotics block the crosslinking required to maintain the peptidoglycan meshwork, the main component in bacterial cell walls. These antibiotics target penicillin-binding proteins (PBPs), which catalyze the incorporation of new material into the peptidoglycan during bacterial growth. In Gram-negative bacteria, two proteins from this family, PBP2 and PBP3, play essential roles in cell elongation and division, respectively.
Sci Adv. 2020 Feb 12;6(7):eaay4453. eCollection 2020 Feb.
Castañeda-García A, Martín-Blecua I, Cebrián-Sastre E, Chiner-Oms A, Torres-Puente M, Comas I, Blázquez J.
Abstract
The postreplicative mismatch repair (MMR) is an almost ubiquitous DNA repair essential for maintaining genome stability. It has been suggested that Mycobacteria have an alternative MMR in which NucS, an endonuclease with no structural homology to the canonical MMR proteins (MutS/MutL), is the key factor. Here, we analyze the spontaneous mutations accumulated in a neutral manner over thousands of generations by Mycobacterium smegmatis and its MMR-deficient derivative (ΔnucS). The base pair substitution rates per genome per generation are 0.004 and 0.165 for wild type and ΔnucS, respectively. By comparing the activity of different bacterial MMR pathways, we demonstrate that both MutS/L- and NucS-based systems display similar specificity and mutagenesis bias, revealing a functional evolutionary convergence. However, NucS is not able to repair indels in vivo. Our results provide an unparalleled view of how this mycobacterial system works in vivo to maintain genome stability and how it may affect Mycobacterium evolution.
doi: 10.1126/sciadv.aay4453.
Expert Rev Anti Infect Ther. 2020 Feb 13.
Gil-Gil T, Martínez JL, Blanco P.
Abstract
Introduction: Stenotrophomonas maltophilia is a prototype of bacteria intrinsically resistant to antibiotics. The reduced susceptibility of this microorganism to antimicrobials mainly relies on the presence in its chromosome of genes encoding efflux pumps and antibiotic inactivating enzymes. Consequently, the therapeutic options for treating S. maltophilia infections are limited.Areas covered: Known mechanisms of intrinsic, acquired and phenotypic resistance to antibiotics of S. maltophilia and the consequences of such resistance for treating S. maltophilia infections are discussed. Acquisition of some genes, mainly those involved in co-trimoxazole resistance, contributes to acquired resistance. Mutation, mainly in the regulators of chromosomally-encoded antibiotic resistance genes, is a major cause for S. maltophilia acquisition of resistance. The expression of some of these genes is triggered by specific signals or stressors, which can lead to transient phenotypic resistance.Expert opinion: Treatment of S. maltophilia infections is difficult because this organism presents low susceptibility to antibiotics. Besides, it can acquire resistance to antimicrobials currently in use. Particularly problematic is the selection of mutants overexpressing efflux pumps since they present a multidrug resistance phenotype. The use of novel antimicrobials alone or in combination, together with the development of efflux pumps' inhibitors may help in fighting S. maltophilia infections.
doi: 10.1080/14787210.2020.1730178.
- La infección por la bacteria E. coli enterohemorrágica (EHEC) se transmite por alimentos contaminados produciendo colitis hemorrágicas, anemia hemolítica e insuficiencia renal, lo que puede llegar a causar graves secuelas y la muerte.
- No existe en la actualidad vacuna o una terapia eficiente, pero investigadores del CSIC han encontrado una novedosa estrategia que puede bloquear la infección.
La bacteria E. coli enterohemorrágica (EHEC) se encuentra a menudo presente en el intestino del ganado bovino sin causar síntomas, pero desde estos animales puede llegar a contaminar alimentos (p.ej. carne picada, vegetales, zumos) si no se toman las medidas sanitarias adecuadas durante su elaboración. Tras la ingestión de alimentos contaminados EHEC infecta el colon humano y produce toxinas que causan colitis hemorrágica, anemia hemolítica e insuficiencia renal, lo que puede generar graves efectos secundarios e incluso la muerte. Estas infecciones suponen un riesgo para la salud pública ya que no existe vacuna ni una terapia efectiva frente a EHEC, ya que el uso de antibióticos puede inducir una mayor liberación de toxinas.
J Antimicrob Chemother. 2019 Aug 1.
Blanco P, Corona F, Martinez JL.
Abstract
OBJECTIVES: To elucidate the potential mutation-driven mechanisms involved in the acquisition of tigecycline resistance by the opportunistic pathogen Stenotrophomonas maltophilia. The mutational trajectories and their effects on bacterial fitness, as well as cross-resistance and/or collateral susceptibility to other antibiotics, were also addressed.
METHODS: S. maltophilia populations were submitted to experimental evolution in the presence of increasing concentrations of tigecycline for 30 days. The genetic mechanisms involved in the acquisition of tigecycline resistance were determined by WGS. Resistance was evaluated by performing MIC assays. Fitness of the evolved populations and individual clones was assessed by measurement of the maximum growth rates.
RESULTS: All the tigecycline-evolved populations attained high-level resistance to tigecycline following different mutational trajectories, yet with some common elements. Among the mechanisms involved in low susceptibility to tigecycline, mutations in the SmeDEF efflux pump negative regulator smeT, changes in proteins involved in the biogenesis of the ribosome and modifications in the LPS biosynthesis pathway seem to play a major role. Besides tigecycline resistance, the evolved populations presented cross-resistance to other antibiotics, such as aztreonam and quinolones, and they were hypersusceptible to fosfomycin, suggesting a possible combination treatment. Further, we found that the selected resistance mechanisms impose a relevant fitness cost when bacteria grow in the absence of antibiotic.
CONCLUSIONS: Mutational resistance to tigecycline was easily selected during exposure to this antibiotic. However, the fitness cost may compromise the maintenance of S. maltophilia tigecycline-resistant populations in the absence of antibiotic.
doi: 10.1093/jac/dkz326
COOKIES POLICY
A cookie is a text file that is stored on your computer or mobile device via a web server and only that server will be able to retrieve or read the contents of the cookie and allow the Web site remember browser preferences and navigate efficiently. Cookies make the interaction between the user and the website faster and easier.
General information
This Website uses cookies. Cookies are small text files generated by the web pages you visit, which contain the session data that can be useful later in the website. In this way this Web remembers information about your visit, which can facilitate your next visit and make the website more useful.
How do cookies?
Cookies can only store text, usually always anonymous and encrypted. No personal information is ever stored in a cookie, or can be associated with identified or identifiable person.
The data allow this website to keep your information between the pages, and also to discuss how to interact with the website. Cookies are safe because they can only store information that is put there by the browser, which is information the user entered in the browser or included in the page request. You can not run the code and can not be used to access your computer. If a website encrypts cookie data, only the website can read the information.
What types of cookies used?
The cookies used by this website can be distinguished by the following criteria:
1. Types of cookies as the entity that manages:
Depending on who the entity operating the computer or domain where cookies are sent and treat the data obtained, we can distinguish:
- Own cookies: are those that are sent to the user's terminal equipment from a computer or domain managed by the editor itself and from which provides the service requested by the user.
- Third party cookies: these are those that are sent to the user's terminal equipment from a machine or domain that is not managed by the publisher, but by another entity data is obtained through cookies.
In the event that the cookies are installed from a computer or domain managed by the editor itself but the information collected by these is managed by a third party can not be considered as party cookies.
2. Types of cookies as the length of time that remain active:
Depending on the length of time that remain active in the terminal equipment can be distinguished:
- Session cookies: cookies are a type designed to collect and store data while the user accesses a web page. Are usually used to store information that only worth preserving for the service requested by the user at any one time (eg a list of products purchased).
- Persistent cookies: cookies are a type of data which are stored in the terminal and can be accessed and treated for a period defined by the head of the cookie, and can range from a few minutes to several years.
3. Cookies types according to their purpose:
Depending on the purpose for which the data are processed through cookies, we can distinguish between:
- Technical cookies: these are those that allow the user to navigate through a web page or application platform and the use of different options or services it exist as, for example, control traffic and data communication, identify the session, access to restricted access parts, remember the elements of an order, make the buying process an order, make an application for registration or participation in an event, use security features while browsing store content for dissemination videos or sound or share content via social networks.
- Customization cookies: these are those that allow the user to access the service with some general characteristics based on a predefined set of criteria in the user terminal would eg language, the type of browser through which you access the service, the locale from which you access the service, etc.
- Analysis cookies: they are those that allow the responsible for them, monitoring and analyzing the behavior of users of the web sites that are linked. The information gathered through such cookies are used in measuring the activity of web sites, application or platform and for the profiling of user navigation of such sites, applications and platforms, in order to make improvements function data analysis how users use the service.
Management tool cookies
This Website uses Google Analytics.
Google Analytics is a free tool from Google that primarily allows website owners know how users interact with your website. Also, enable cookies in the domain of the site in which you are and uses a set of cookies called "__utma" and "__utmz" to collect information anonymously and reporting of website trends without identifying individual users..
For statistics of use of this website use cookies in order to know the level of recurrence of our visitors and more interesting content. This way we can concentrate our efforts on improving the most visited areas and make the user more easily find what they are looking for. On this site you can use the information from your visit for statistical evaluations and calculations anonymous data and to ensure the continuity of service or to make improvements to their websites. For more details, see the link below privacy policy [http://www.google.com/intl/en/policies/privacy/]
How to manage cookies on your computer: disabling and deleting cookies
All Internet browsers allow you to limit the behavior of a cookie or disable cookies within settings or browser settings. The steps for doing so are different for each browser, you can find instructions in the help menu of your browser.
If you decline the use of cookies, since it is possible thanks to the preferences menu of your browser or settings, reject, this website will continue to function properly without the use of the same.
Can you allow, block or delete cookies installed on your computer by setting your browser options installed on your computer:
- For more information about Internet Explorer click here.
- For more information on Chrome click here.
- For more information about Safari click here.
- For more information about Firefox click here.
Through your browser, you can also view the cookies that are on your computer, and delete them as you see fit. Cookies are text files, you can open and read the contents. The data within them is almost always encrypted with a numeric key corresponding to an Internet session so often has no meaning beyond the website who wrote it.
Informed consent
The use of this website on the other hand, implies that you paid your specific consent to the use of cookies, on the terms and conditions provided in this Cookies Policy, without prejudice to the measures of deactivation and removal of cookies that you can take, and mentioned in the previous section.