Juan Nogales
Group Leader

Research summary
Our goal is deciphering the complexity of microbial metabolism, its evolutionary and biotechnological implications through a multidisciplinary approach. By assuming that the whole is greater than the sum of its parts, we aim to contribute to a better understanding of the emergent properties of microbial systems at subcellular, cellular and supracellular levels. We pursuit the rational re-design of these system properties towards novel biotechnological and medical applications.
Publications
San León D, and Nogales J. Towards merging Bottom-up and Top-down model-based designing of synthetic microbial communities. Current Opin in Microbiol. 2022 69: 102169
Manoli MT, Nogales J, and Prieto A. Synthetic control of metabolic states in Pseudomonas putida by tuning polyhydroxyalkanoate cycle. Mbio 2022 13 (1), e01794-21
Nogales J and Garmendia J. Bacterial metabolism and pathogenesis intimate intertwining: time for metabolic modelling to come into action Microb Biotechnol. 2021 Oct 21.doi: 10.1111/1751-7915.13942.
Gudmundsson S and Nogales J. Recent advances in model-assisted metabolic engineering. Curr Opin Syst Biol. 2021; Vol 28,100392
Torres-Bacete J, García JL, and Nogales J. A portable library of phosphate-depletion based synthetic promoters for customable and automata control of gene expression in bacteria. Microb Biotechnol. 2021 Mar 30.doi: 10.1111/1751-7915.13808.
García-Jiménez B, Torres-Bacete J, Nogales J. Metabolic modelling approaches for describing and engineering microbial communities. Comput. Struct. Biotechnol. J. 2021. 19, 226-246
Funding
 2017-2024: INFRAIA-02-2017 and H2020-INFRADEV-2019-2. Industrial Biotechnology Innovation and Synthetic Biology Accelerator, IBISBA nº GA 730976 and 871118.
2017-2024: INFRAIA-02-2017 and H2020-INFRADEV-2019-2. Industrial Biotechnology Innovation and Synthetic Biology Accelerator, IBISBA nº GA 730976 and 871118.
2019-2023: H2020-NMBP-BIO-2018-two-stage. Synthetic microbial consortia-based platform for flavonoids production using synthetic biology, SynBio4Flav nº GA 814650.
2020-2023: ERA CoBioTech, 2nd Joint Call on Biotechnologies. Microbial Conversion of lignin to monomers for bio-based plastic using synthetic biology MILIMO.
2020-2024: H2020-NMBP-BIO-CN-2019. MIXed plastics biodegradation and UPcycling using microbial communities, MIXUP nº GA 870294.
2020-2023: MCIU. System analysis and biotechnological applications of bacterial metabolic robustness at supracellular level (RobExplode). PID2019-108458RB-I00.
2021-2025: H2020-FNR-2020-2. Harnessing the power of nature through productive microbial consortia in biotechnology: Measure, Model, Master, PROMICOM , nº GA 101000733

Interdisciplinary Platform for Sustainable Plastics towards a Circular Economy‐Spanish National Research Council (SusPlast-CSIC).
Our foundational aim is the system-level understanding of microbial metabolism as a framework for developing a broad range of novel and non-intuitive biotechnological processes. Taking advantage of metabolic modelling, systems and synthetic biology we are addressing, at different levels, the understanding and full taming of bacterial systems emergence.
Increasing the completeness and scope of metabolic reconstructions
We are involved in the high-quality metabolic modeling of a large set of metabolically diverse bacteria including P. putida, S. elongatus, A. platensis, Azoarcus CIB, S. granuli, P. pseudoalcaligenes, B. bacteriovorus, H. influenzae and Bifidubacterium sps. This effort is enabling the system-level analysis of new metabolic processes while providing new computational test-beds for biotechnological applications. We are particularly interested in the inclusion of new metabolic modules such as the generation of reactive oxygen species, underground metabolisms and metabolic heterogeneity.
System-level analysis of Metabolic Robustness in bacteria
The robustness of a system is the property that allows it to maintain its functions despite perturbations. Through the metabolic modeling analysis of P. putida, we have identified metabolic cycles providing robustness. The synthetic biology assisted validation of such cycles is allowing the rational engineering of superior microbial biocatalyst under diverse biotechnological scenarios.
System-level analysis and designing of microbial communities
The division of labor allows an expanded complexity and functionality in bacteria. We are interested in: i) understanding how these expanded capabilities emerge within a bacterial populations and communities and ii) how we can engineer this supracellular-level functionality towards biotechnological endeavors. To address these two fundamental questions, we have developed systems and synthetic biology tools for modeling and engineering synthetic microbial populations and consortia. We are applying this technology in the revalorization of complex polymers such as lignin and plastic waste as well as in the cost effective production of plant-based secondary metabolites such as flavonoids.
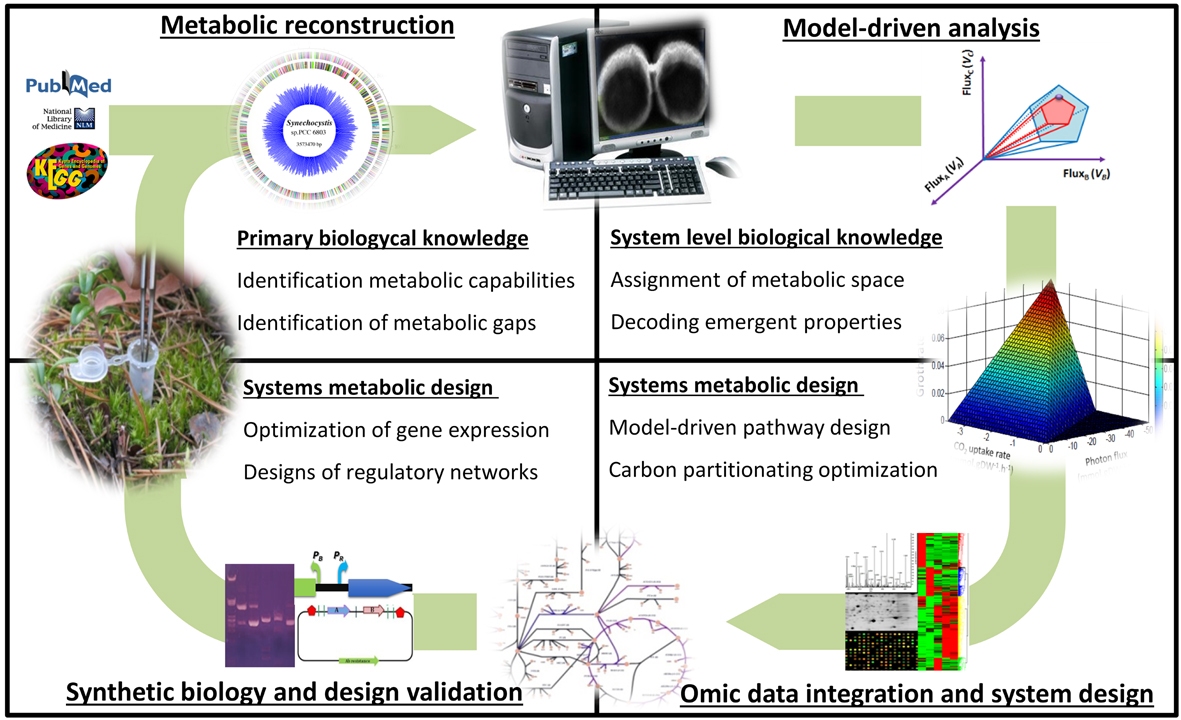
Systems metabolic modeling and engineering platform developed by SBG.
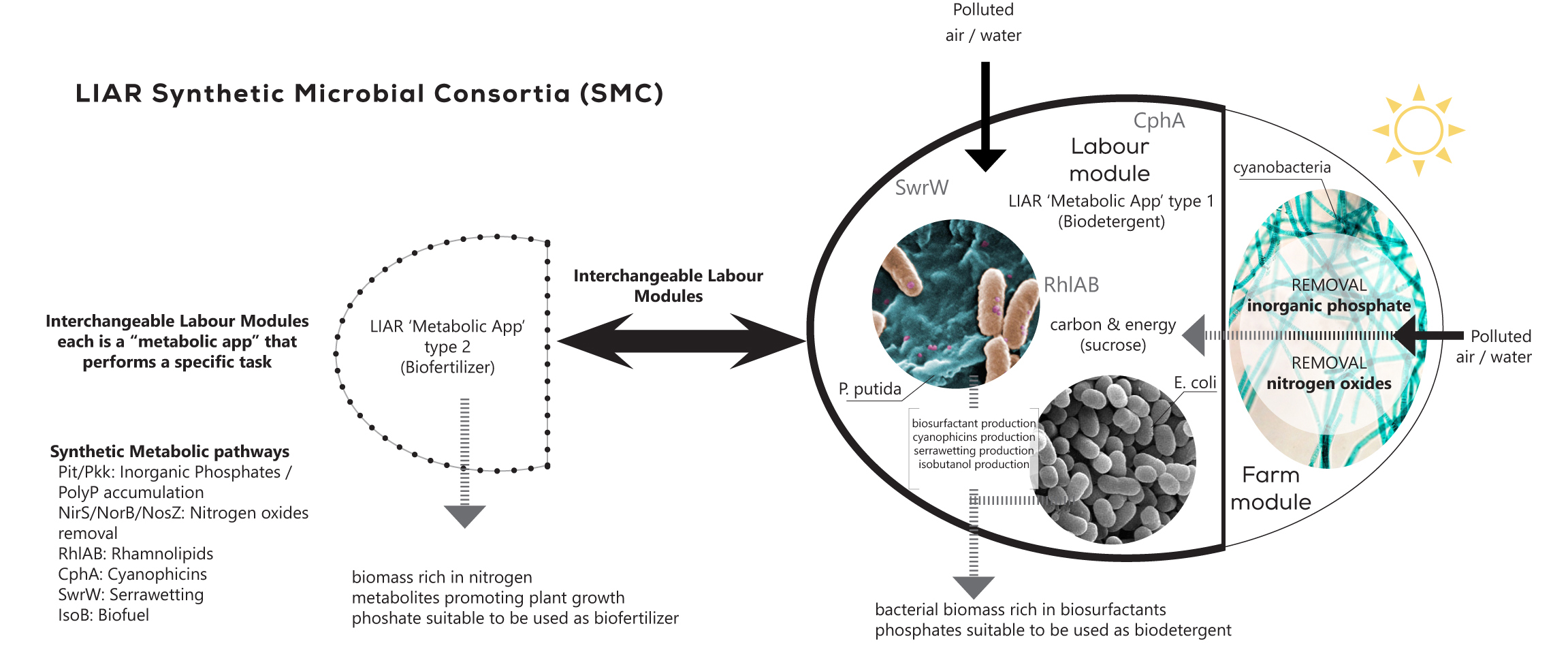
Multitasks synthetic microbial consortia being developed by SBG in the context of Living Architecture project http://livingarchitecture-h2020.eu/








