Contact: José Requejo-Isidro
Single-molecule investigation of molecular events at the membrane
The interaction between a protein and the membrane depends not only on the particular chemical species involved in the interaction, but also on collective biophysical properties of the membrane such as surface potential, rigidity or curvature. The nature of this interaction (lipid-specific, electrostatic or driven by the hydrophobic effect), determines, in turn, the diffusional regime of the protein at the membrane. Therefore, the correlation of protein diffusion with lipid composition and membrane biophysical properties provides us with a way to interrogate lipid-protein interactions. At CNB, we use single-molecule tracking (SMT) to measure investigate lipid-protein interaction at the single-molecule level.
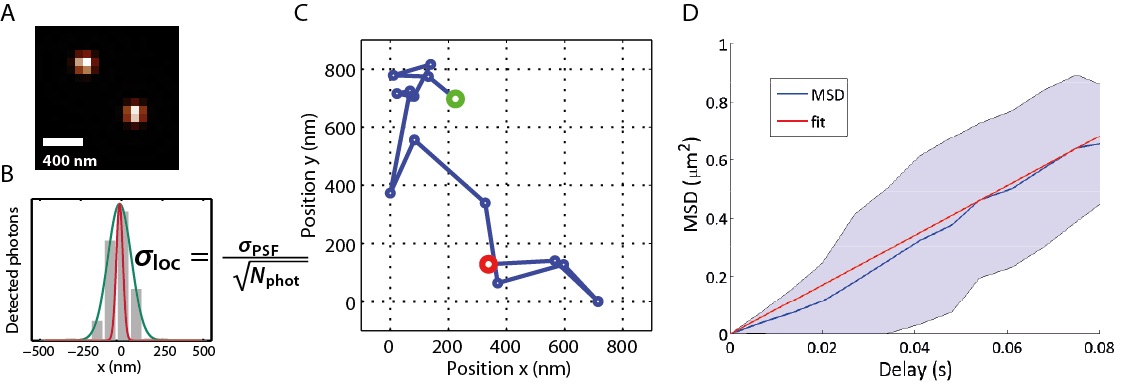
Single-Molecule Tracking (SMT): (A) Two spatially isolated molecules imaged through an optical microscope. (B) The localisation accuracy, σloc (in red), is dependent on the number of photons that have been detected (grey) and is much higher than that obtained from the emission pattern of the emitter (green). In our lab single molecules are routinely localised within 30 nm accuracy. (C) Track of a single Annexin V molecule diffusing on a supported lipid bilayer. (D) The Mean-Squared Displacement (MSD) of the Annexin molecule in (C) is linear with diffusion time, indicating brownian diffusion.
Single-molecule investigation of inter- and intramolecular protein dynamics
One suitable way to investigate inter and intramolecular protein interactions is FRET (Förster Resonance Energy Transfer). FRET may occur when two fluorescent molecules are closer than 10 nm, which is commonly the signature of interaction of proteins (intermolecular FRET) or protein domains (intramolecular FRET). However, transient conformers that give rise to a short-lived FRET signal may be outnumbered by more stable molecular states with a longer, well defined, FRET signal. In addition, several interacting geometries may be possible, all resulting in similar, but slightly different FRET signatures. Ensemble FRET measurements average out all signals, favouring those interacting geometries or conformers with the strongest FRET. As a consequence, information on transient or less-probable molecular arrangements is lost. On the other hand, observation of individual molecule behaviour allows identification of those molecular states that yield weaker FRET. At CNB, we are setting up a high-throughput TIRF microscope with single-molecule detection capabilities to measure FRET at the single-molecule level (smFRET).
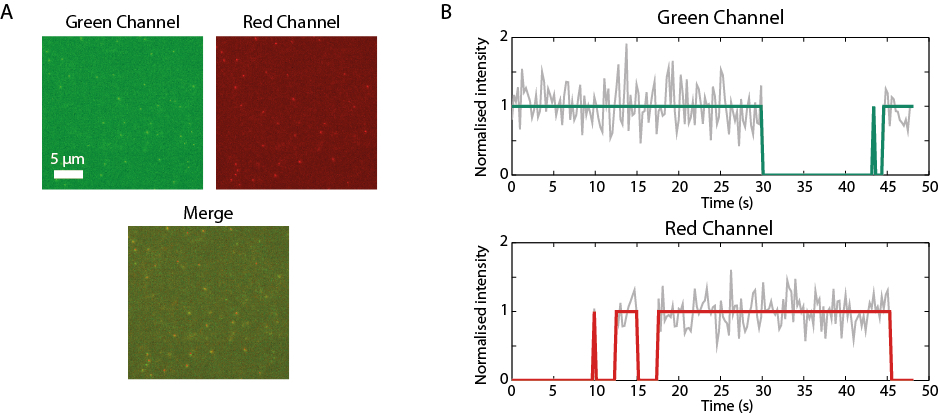
Single-molecule FRET (smFRET): (A) Single-molecule images of green- and red- tagged fluorescent molecules acquired on a TIRF microscope. The green molecules are immobilised on a coverslip, while the red molecules freely diffuse in solution. (B) Information on binding kinetics and transition rates is obtained through correlative analysis of the emission of individual molecules in both microscope channels.
Correlative force of molecular interactions and spatio-temporal dynamics
Correlative Optical Tweezers – Fluorescence Microscopy (CTFM) is a single-molecule technique that combines optical tweezers, fluorescence microscopy and microfluidics. It allows to simultaneously correlate molecular mechanical properties to spatial and temporal dynamics of molecular events. In particular, at CNB we are interested on fundamental aspects of protein- DNA interactions and chromatin organization during cell division.





