Role Of Ubiquitin In The Control Of Plant Growth And Stress Tolerance
RESEARCH GROUP

Vicente Rubio Muñoz
Group Leader
Research Summary
Plant ubiquitination machineries have been proposed to act as hubs to integrate signals from different abiotic stresses and environmental stimuli in order to trigger coherent adaptive and developmental responses in plants. Among them, CRL4-CDDD E3 ubiquitin ligases are key regulators of plant responses to different environmental cues, such as light conditions variation and abiotic stresses, acting as repressors during signalling of the so-called water stress hormone abscisic acid (ABA). Based on previous findings resulting from our long-lasting interest on the functional characterization of the plant proteostasis system, we propose that CRL4-CDDD act as protein-protein interaction platforms that, not only promote ubiquitination and proteasomal degradation of specific protein targets, but also bring together different chromatin-associated activities to coordinately regulate their function according to changing ambient conditions.
Research Lines
To ensure their survival, plants need to develop efficient signal transduction pathways that confer them the ability to respond to sudden changes in environmental cues effectively. These signal transduction pathways often rely on tight control of protein accumulation levels as well as on transcriptional regulation of key genes. In the first case, protein degradation can be controlled by the post-translational modification (PTM) ubiquitination, catalysed by different families of E3 ubiquitin (Ub) ligases, and the subsequent proteasome-mediated degradation of the substrate. Among E3 ligases, CULLIN 4 RING E3 Ub ligases (CRL4) play a pivotal role in modulating protein levels affecting a myriad of plant responses including embryogenesis, seedling photomorphogenesis, circadian clock function, flowering and tolerance to different stresses.
Plants also integrate and respond to environmental cues by chromatin remodelling, which allows for rapid and large-scale reprogramming of gene expression. As eukaryotes, plants have their genome packed within nucleosomes, consisting in ∼147bp of DNA wrapped around protein octamers composed of histones H2A, H2B, H3 and H4, while H1 acts as a linker histone. Albeit mostly evolutionary conserved, genetic amplifications in Arabidopsis have led to the presence of multiple histone variants with specific functions which can displace their canonical counterparts in specific cases. Concomitantly, histone positioning, gene occupancy or functional histone properties, all influencing DNA accessibility, can be altered via covalent PTM of histones. Histone PTM are catalysed by enzymes known as histone writers and erasers, which are capable of laying a wide array of chemical reactions in the flexible areas known as histone tails (both N- and C-terminal tails). Common histone PTMs include methylation, ubiquitination and acetylation.
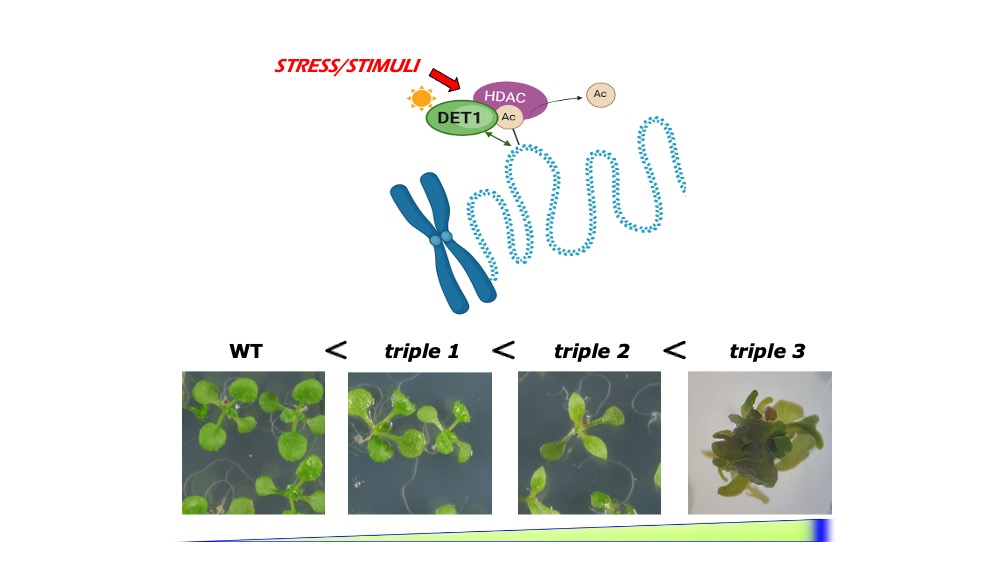
DET1 associates with chromatin remodellers, such as histone deacetylases (HDAC), whose mutation can cause severe developmental defects.
Previous results from our group indicate that CRL4 ubiquitin ligases are key elements in the interconnection between plant proteostasis and control of chromatin remodelling, although their functional relevance is starting to be recognized. To widen our knowledge on CRL4-CDDD-mediated control of chromatin remodeling and transcriptional reprogramming in response to specific cues (i.e. ABA-mediated stresses and light information). Our research interests are summarized in the following General Objective: to characterize new molecular mechanisms, acting at the nexus between protein ubiquitination and degradation, chromatin remodeling and transcriptional control, by which CRL4-CDDD allow integration of environmental signals that trigger specific plant developmental and adaptive responses to environmental stresses, using as model Arabidopsis and oilseed rape plants.
To cope with this objective, our research is carried out within the frame of “EpiSeedLink”, a pan-european consortium, involving 11 partners both from academia and industry, with a special interest in understanding the role of chromatin dynamics in the establishment of plant developmental programs in early stages of their life, and how to translate their discoveries into biotechnological products or applications. “EpiSeedLink” has been granted a Marie Skłodowska-Curie Doctoral Network (MSCA-DN) action (Call 2021), and is coordinated by Vicente Rubio from the CNB-CSIC. The main objective of this proposal is to train the next generation of researchers who will fully exploit the potential of plant epigenetics to address societal challenges, such as ensuring food quality and supply under a likely climate change scenario.
Publications
Members
Staff Scientist
Cristina Martínez
PhD candidates
Martín Albacete
Paolo Lucas Rodrigues
Daniel García

Funding
Our research is funded by national and international institutions as indicated below. For more details, please check the general Funding Section at the CNB website.



News
Identificado el mecanismo que controla la actividad de una hormona vegetal esencial en la respuesta a la sequía
14 de noviembre 2024 En plantas, la mayoría de procesos biológicos están finamente regulados por hormonas vegetales El ácido abscísico (ABA) desempeña un papel fundamental en respuesta a la sequía, la salinidad y las temperaturas extremas Un equipo de investigación...



