Plant-Pathogen Interaction in Viral Infections
RESEARCH GROUPS

Juan Antonio García
Group Leader

Carmen Simón
Group Leader

Adrian A. Valli
Group Leader
Research Summary
The negative impact of pathogens on crop yield is highly concerning, with losses accounting for approximately 30% of global productivity. In our laboratory, we focus on viral plant pathogens with two primary objectives: understanding how they cause disease and identifying networks of plant proteins that act as defensive barriers. To achieve these goals, we employ a multidisciplinary approach that encompasses synthetic biology, various omics techniques, structural studies, and viral ecology among others. The outcomes of our research not only deepen our knowledge about plant-virus interaction and coevolution but also empower us to develop more effective strategies for enhancing antiviral resistance in plants.
Research Lines
Food security is under threat due to the rising global population and the adverse effects of global warming on crop production. In this context, the negative impact of pathogens on crop yield is particularly concerning, with losses amounting to roughly 30% of global productivity. While plants in nature are often infected by viruses that cause no symptoms or even result in mutualistic associations, plant viruses can also induce severe diseases. Breeding for resistance has been effective in combating some viral diseases, but natural sources of resistance are scarce. Therefore, understanding natural resistance mechanisms and viral infection processes is crucial for identifying appropriate targets and advancing biotechnological applications aimed at protecting plants from viral infections.
For our studies we primarily focus on members of the Potyviridae family, the most numerous and socio-economically relevant plant RNA viruses. Over the last years, part of our team has focused its attention on two viral functions that have not been extensively studied by using plum pox virus (PPV, which causes the devastating disease named sharka in prunus trees) as model: the proteolytic processing of viral polyproteins and the post-translational modifications (PTMs) of viral proteins. We have demonstrated that the efficiency of the potyviral leader protease may be carefully regulated to prevent the uncontrolled release of the silencing suppressor HCpro, which could trigger antiviral defenses through complex hormonal and transcriptomic changes. Additionally, our research suggests that alterations in the proteolytic cleavage between NIapro and VPg proteins may play a role in the unique escape of PPV from the HR-like resistance observed in certain Prunus domestica cultivars. Regarding PTMs, our findings indicate that the combined and opposing actions of O-GlcNAcylation and phosphorylation at the N-terminal protrusion of the PPV capsid protein regulate its stability, while phosphorylation at the core region controls the assembly and disassembly of viral particles.

Unraveling the mechanism of induction of hypersensitive response associated to resistance to Plum pox virus in European plums. (a) Resistant Prunus domestica trees inoculated with standard PPV-D isolate, the resistance-escaping isolate PPV-DH or a chimeric virus carrying the PPV-D background with the NIa sequence of PPV-DH. Schematic representation of each virus can be seen on the bottom of each picture. (b) In planta expression of NIa proteins from PPV-D and PPV-DH with a Myc tag to detect cleavage activity by western blot. Quantification of the cleavage percentage observed for each protease is shown in the panel.
Another part of our team is focused on understanding the role of recently acquired viral genes in two distinct potyvirids: Ugandan cassava brown streak virus (UCBSV) and endive necrotic mosaic virus (ENMV). In the case of UCBSV, we are characterizing a viral-derived pyrophosphatase of non-canonical nucleotides (ITP/XTP), named HAM1, expressed by all living organisms and just a few RNA viruses infecting euphorbiaceous plants in nature. For ENMV, we are investigating the viral P1 protein, which contains an AlkB domain with an unknown function potentially related to RNA methylation. We hypothesize that these viruses have evolved to acquire new genes to counteract plant antiviral mechanisms, such as the overaccumulation of non-canonical nucleotides and RNA methylation. In addition, we have recently begun exploring the three-trophic interaction between plants, plant viruses, and plant-associated microbiota. We believe that commensal microbes interacting with plants may play a significant role in modulating plant defenses against pathogenic viruses in natural ecosystems.
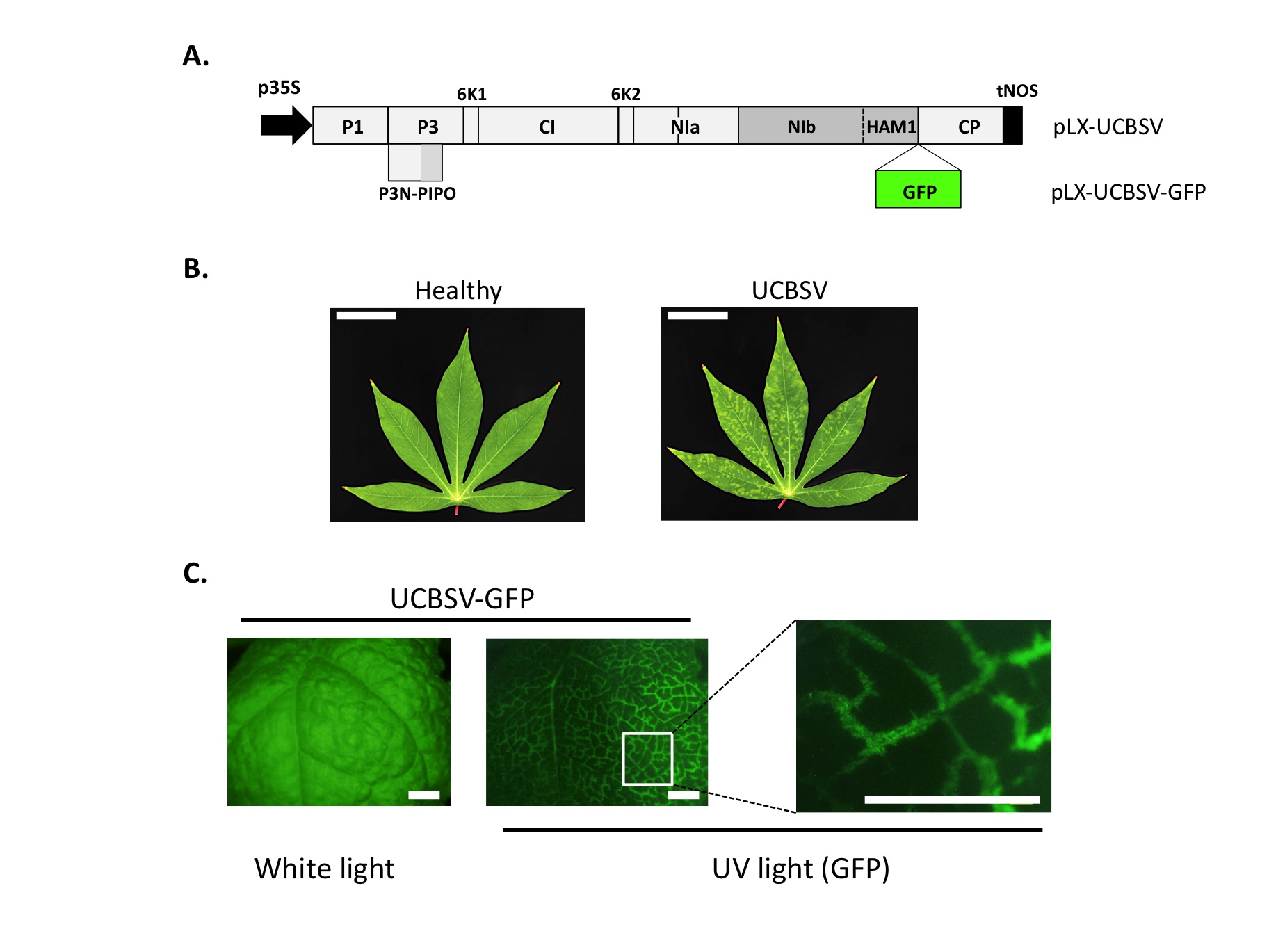
Infection of plants with laboratory-designed cDNA clones of UCBSV. (A) The RNA genome of UCBSV was retrotranscribed to cDNA and this sequence was then inserted in between a strong promoter (p35S from Cauliflower mosaic virus) and a terminator (tNOS from Agrobacterium nopaline synthase) to generate the infectious cDNA plasmid termed pLX-UCBSV (Pasin et al., 2017, ACS Synthetic Biology). This plasmid was further modified to tag the virus with GFP, thus generating pLX-UCBSV-GFP. (B) Leaves of cassava plants (Manihot esculenta) at 2 months post-inoculation observed under white light (bar = 4 cm). Symptoms of infection are clearly detected in upper leaves of plants inoculated with pLX-UCBSV. (C) One leaf of a Nicotiana benthamiana plant, infected with UCBSV-GFP, observed under white light and UV light (to detect GFP-derived fluorescence) with a stereomicroscope. GFP is distributed across primary and secondary veins as the virus spread through the leaf (bar = 0.5 cm).
Publications
Group Members

Group Leaders
Juan Antonio García
Carmen Simón
Adrian A. Valli
Staff scientists
Beatriz García García
Sandra Martínez Turiño
Bernardo Rodamilans
Technicians
Irene Gonzalo
Postdoctoral scientists
Alberto Cobos
Mario Ledesma
Mario Rincón
PhD candidates
Rafael García López
Julio Aragón Lago
Funding
News
El CNB producirá tejidos vegetales artificiales para estudiar infecciones virales
22 de octubre 2024 Un nuevo proyecto financiado por la Organización Internacional Human Frontier Science Program codirigido por investigadores del CSIC desarrollará una plataforma para producir tejidos artificiales en 3D La convocatoria de ayudas “Research Grants...
Redefinen el mecanismo y la relevancia del “patinaje” de polimerasas de virus ARN
18 de Septiembre 2024 Las polimerasas son proteínas encargadas de copiar el material genético de los organismos, tanto ADN como ARN Los virus pequeños pueden optimizar la expresión de más genes utilizando el “patinaje de la polimerasa”, un mecanismo que permite...
Juan Antonio García y Jorge Cuellar reciben la Medalla Margarita Salas del CSIC a la supervisión de tesis doctorales
3 de Julio 2024 El Consejo Superior de Investigaciones Científicas (CSIC) ha celebrado esta semana el acto de entrega de los Premios a la Supervisión Novel, los Premios de Tesis Doctorales Relevantes del CSIC y la Medalla Margarita Salas a la mejor trayectoria en...






