Biocomputing
RESEARCH GROUPS

Carlos Oscar S. Sorzano
Group Leader
Research Summary
We work in creating the next generation of data processing and management software and algorithms in cryo Electron Microscopy, both for the work on isolated macromolecules and for in situ cellular Electron Microscopy.
Research Lines
Our group develops the next generation of enabling integrative structural biology technologies. We focus on new algorithms and new software for the analysis of molecular and atomic resolution images acquired by an Electron Microscope under cryogenic conditions. The goal is to help understanding biological function through structural information, including macromolecular flexibility. The image processing algorithms we are developing cover the whole pipeline from the microscope to the final structure. We work on algorithms that are applied to images coming from purified samples following what is referred as Single Particle Analysis, and also on new functionalities in the quickly expanding area of Electron Tomography, aiming at obtaining structural information in the context of the whole cell, without potential disturbance coming from purification.
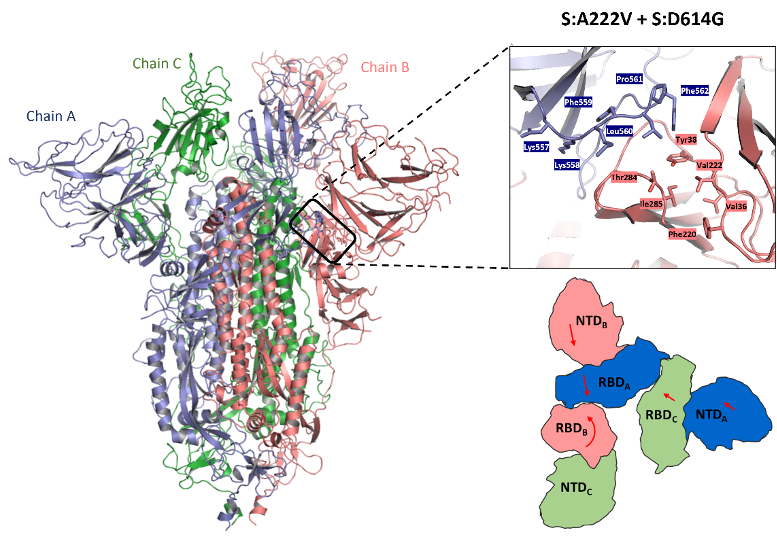
Effect of Spanish variant spike mutation A222V from Cryo-EM: single particle analysis resulted in a single dominant class average structure with 1 receptor binding domain (RBD) in the open/up state ready to bind its host receptor ACE2. The mutation lies at an interface between the NTD to which it belongs and the neighbouring hinge region at the base of the RBD (zoomed), leading to small rearrangements in the packing of the NTDs and RBDs as shown in the schematic. This ultimately impacted its flexibility as shown in B-factors and molecular dynamics (not shown).
All or software and algorithms behind are open-source and publicly available under the software package Xmipp. We also develop a workflow engine especially well-suited to image processing in Structural Biology called Scipion that integrates Xmipp and many other software suites. Scipion provides a traceability, reproducibility and interoperability layer on top of the scientific software suites that integrate. We are particularly committed to Open Science and to provide the software tools that allow the FAIR use of the electron microscopy data (FAIR=Findable, Accessible, Interoperable, and Reusable). Once the structure of a macromolecule is solved, our tools allow going further by helping to construct an atomic model and to explore possible ligands that could interact with it. Finally, we also have a role in Structural Bioinformatics by integrating genomic, proteomic, and interactomic information onto the solved structure. Our tool in this area is 3DBionotes, and it has been recognized as one of the few recommended Interoperability Resources of the European Infrastructure of Life Science Information, ELIXIR. Finally, part of our group uses the image processing tools developed by us to solve specific structures. In this way, we know first-hand the problems encountered in the leading edge of Structural Biology.
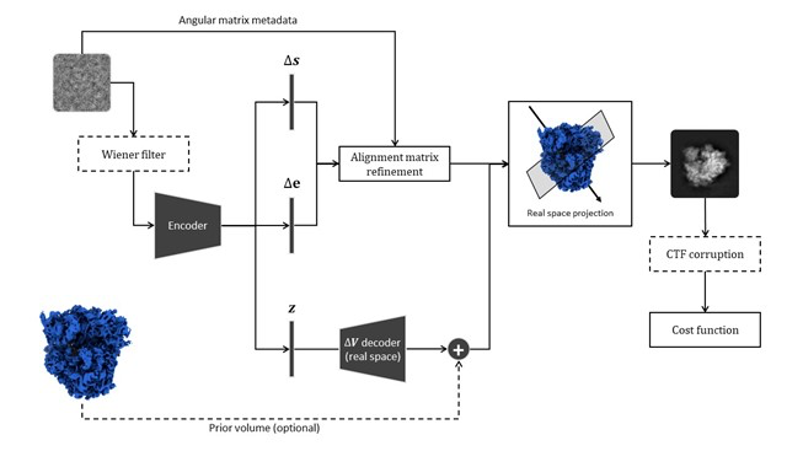
Basic network architecture for conformational and compositional analysis of cryo Electron Microscopy images.
Our group also provides access to dedicated and specialized image processing to the whole of the European scientific community through the Instruct-ERIC Image Processing Center (I2PC), which part of the Spanish contribution to the European infrastructure for Integrative Structural Biology Instruct-ERIC. I2PC supports the community by having an access program by which we actively help users solving the structures of the macromolecules of their interest, hosting short-term internships, and by a training program through which we give at least 4 technological courses per year on image processing in Electron Microscopy.
Publications
More Publications
Group Members

Group Leaders
José-María Carazo
Carlos Oscar Sánchez Sorzano
Lab Manager
Blanca E. Benítez
Lab assistants
Mikel Iceta
Alberto García Mena
Irene Sanchez Lopez
Jorge Jiménez de la Morera
Mª Dolores Sanchez de Lara
Pablo Conesa Mingo
Oscar Saiz
Staff Scientists
Mª Marta Martínez
Roberto Melero
Erney Ramirez Aportela
Yunior Fonseca Reyna
Claudia Nicole Larriux
Postdoctoral researchers
Marcos Gragera
Jose Luis Vilas
James M. Krieger
David Herreros
PhD candidates
Oier Lauzirika
Daniel Marchan
Daniel del Hoyo
Eduardo García
Funding



News
Las Sinergias Severo Ochoa financiarán 8 proyectos colaborativos de grupos del CNB
La convocatoria interna Sinergias Severo Ochoa del Centro Nacional de Biotecnología (CNB-CSIC) financiará ocho proyectos colaborativos de grupos de investigación del centro, impulsando el desarrollo de programas transversales de alta calidad científica. Entre las...
El CNB-CSIC recibe una ayuda dentro del programa Marie Skłodowska-Curie para avanzar hacia un futuro sostenible
4 de abril 2024 Las becas del programa europeo Allies abordan la resolución de desafíos en inteligencia artificial, robótica y ciencia de datos que contribuya a la consecución de los Objetivos de Desarrollo Sostenible de Naciones Unidas Los recientes avances de...
Launch of ELIXIR-STEERS – enhancing research software management in life science
8 de Febrero 2024 ELIXIR-ES is pleased to announce participation in ELIXIR-STEERS, a three-year EU-funded project with a total budget of €4M, which celebrates its kick-off meeting this week in Brussels (Belgium). The project is a collaboration between all ELIXIR...










