Molecular Infection Biology
RESEARCH GROUPS

Daniel López Serrano
Group Leader
Research Summary
The Molecular Infection Biology (MIB) Lab explores the organizational complexity of bacterial cells, focusing on how bacteria organize and compartmentalize their cellular processes to enhance efficiency and preserve cellular integrity. By understanding these mechanisms, we aim to develop innovative, non-traditional strategies to combat antibiotic-resistant infections. Our research particularly investigates the existence of Functional Membrane Microdomains (FMM) in bacteria, analogous to the lipid rafts in eukaryotic cells, and their role in maintaining proteins correctly folded without the need for ATP. FMM are key structures for preserving the bacterial proteome during infection, and disrupting FMM compromises bacterial viability, leading to attenuated infections. At the MIB Lab, we combine molecular approaches with advanced imaging techniques, such as cryo-electron microscopy, to reveal the structural and functional complexity of FMM-associated cellular processes and to develop anti-FMM drugs to fight bacterial infections that are resistant to conventional antibiotics.
Research Lines
The Molecular Infection Biology MIB lab pioneered the study of Functional Membrane Microdomains (FMM) in bacteria and their role in pathogenesis We and other groups contributed to the field by proving that bacteria organized functional membrane microdomains (FMM) and visualized the assembly of FMM in bacterial membranes using a combination of electron tomography and super-resolution microscopy. In the MIB Lab, we use the human pathogen Methicillin-Resistant Staphylococcus aureus (MRSA) as a model. Our preliminary data showed a severe reduction in virulence and b-lactam resistance of the FMM-defective mutant in murine infection models, which was caused by a decrease in bacterial survival due to a general malfunction of the FMM client proteins, resulting in MRSA infections susceptible to classical b-lactams.

Although FMM play a crucial role in maintaining bacterial virulence and antibiotic resistance, their contribution to preserving protein activity was previously unknown. However, we discovered that bacterial FMM help maintain cellular proteostasis under stress by stabilizing unfolded proteins. Stress causes a portion of the bacterial proteome to unfold, exposing hydrophobic residues that lead to protein aggregation. FMM hydrophobic environment accumulates and stabilizes these proteins, preventing cytotoxic interactions. Flotillin, a key FMM protein, assembles in a clamp-shaped conformation with hydrophobic tentacles that bind and stabilize unfolded proteins, facilitating their refolding at no ATP cost to the cell. This process prevents misfolding-related dysfunction without requiring ATP. In sum, FMM organizes a conserved ATP-independent mechanism dedicated to prevent protein-misfolding bacterial dysfunction.
Our research is organized in three main lines of research; a structural, a functional and an applied lines of research to leverage the diverse expertise of the MIB lab to explore FMM biology. Structurally, we use cryo-EM combined with molecular approaches to describe the cellular programs driving the assembly of FMM in biological membranes and how FMM operate to stabilize unfolded proteins. Functionally, we use molecular approaches to functionally describe how FMM operate independently on ATP and the precise physiological contribution of FMM to bacterial proteostasis. Finally, the applied component of innovation involves translating our basic knowledge into European biomedical innovation, by developing anti-FMM strategies to eliminate antibiotic-resistant bacterial infections by making bacteria vulnerable to the immune response and sensitive to antibiotics.
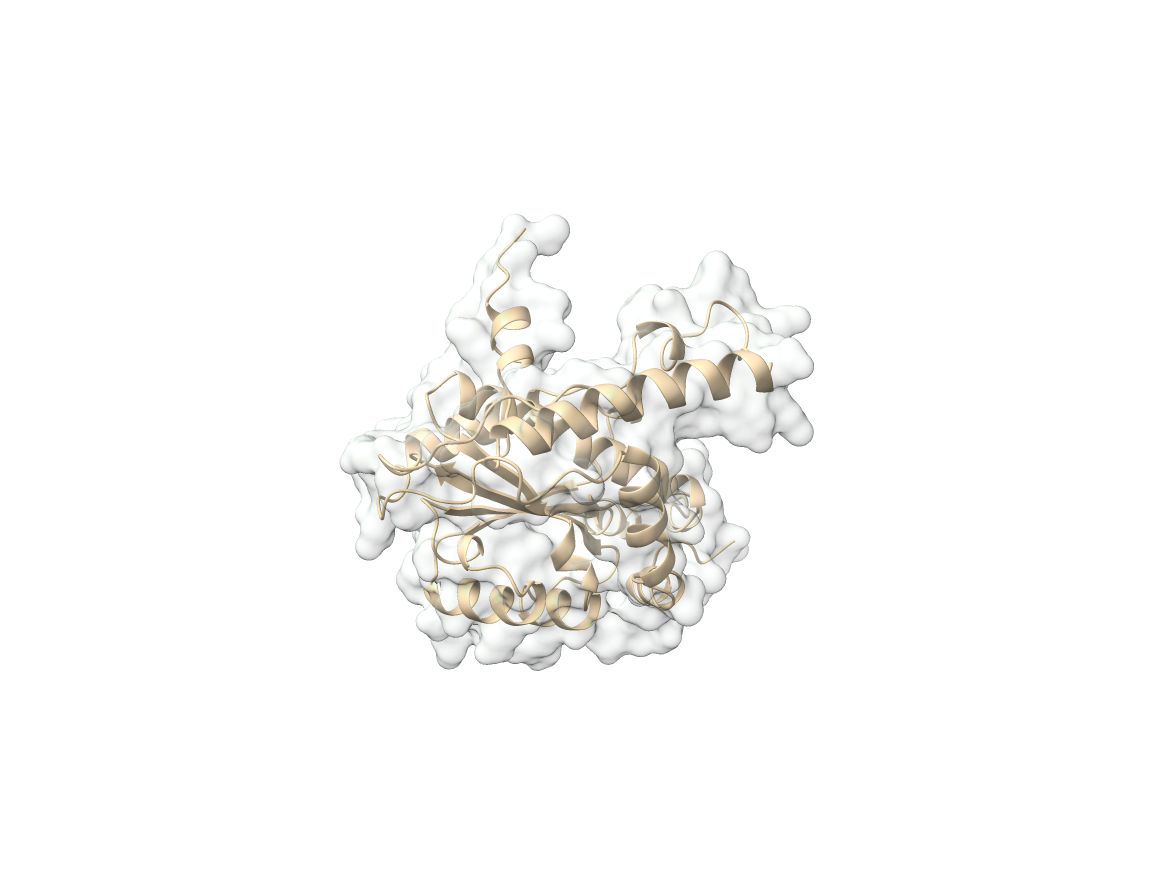
The MIB Lab investigations aim to establish new ground in understanding the cellular programs for compartmentalizing biological membranes and their association with protein-misfolding cellular dysfunction. We will apply this fundamental knowledge to develop anti-FMM drugs that can help to fight MDR bacterial infections and to revitalize currently ineffective antibiotics.
Publications
Ukleja M, Kricks L, Torrens G, Peschiera I, Rodrigues-Lopes I, Krupka M, García-Fernández J, Melero R, del Campo R, Eulalio A, Mateus A, López-Bravo M, Rico AI, Cava F and Lopez D. Flotillin-Mediated Stabilization of Unfolded Proteins in Bacterial Membrane Microdomains Nature Communications 2024, 15:5583. DOI: https://doi.org/10.1038/s41467-024-49951-1
Group Members

Group Leaders
Daniel López Serrano
María López-Bravo
Lab Assistant
Ana Marina Cabrerizo
Project Leaders
Marta Ukleja
Julia García Fernández
Postdoctoral Researchers
Elena Pajares
Esmeralda Solar
PhD candidates
Tamara Alonso Blanco
Héctor Olmeda López
David Torrens González
Marta Matínez López


