Sponge Microbiome
RESEARCH FELLOW

Cristina Diez Vives
Future Leader La Caixa
Contact
Research Summary
Sponges are keystone species in marine environments, essential for the ocean’s health due to their role in both habitat formation and benthic-pelagic coupling. Sponges rely on their associated microbial community for nutrition, health and chemical defence, and hence, are necessary for their survival. In turn, microbes find a stable environment with an abundance of nutrients as a result of their host seawater filtering effort. I study this ancient relationship from different perspectives, and the ecological and evolutionary mechanisms that shape the microbiome complexity and diversity. My reseach is mainly based on molecular tools such as metabarcoding, genomics and transcritomics, and microscopy techniques.
Research Lines
Ecological and evolutionary patterns of sponge symbiosis
The sponge microbiome is fairly consistent in conspecific sponges both spatially and temporally, indicating a very stable and successful host-symbiont association. However larger ecological and evolutionary patterns are underexplored. We aim to uncover whether there are stronger selective forces driving the composition of high microbial abundant sponges than low abundant microbial sponges. Similarly, the deep-sea environment presents environmental constrains that can affect the availability of microbes and, in turn, impact the sponge symbiont composition. Furthermore, we want to assess general patterns of microbiome assemblage in marine vs. freshwater species and among the different sponge reproductive strategies.
The Freshwater Sponge Microbiome Project
Freshwater sponges evolved from the marine species only 32 Mya, forming a monophiletic clade (Spongillida) with around 250 species, compared to the 9000 marine species. Their microbiome was most likely fundamental for this transition, however, it has only been described for a handful of species. Furthermore, the fidelity of the microbiome among same sponge species in freshwater is challenged due to the unconnected habitat they live in. Our goal is to describe how the environment and dispersion mechanisms shape this microbiome, and what other forces have influenced the present microbial composition (i.e. host genome, algal symbionts, glaciation events, etc).
The reproductive sponge microbiome
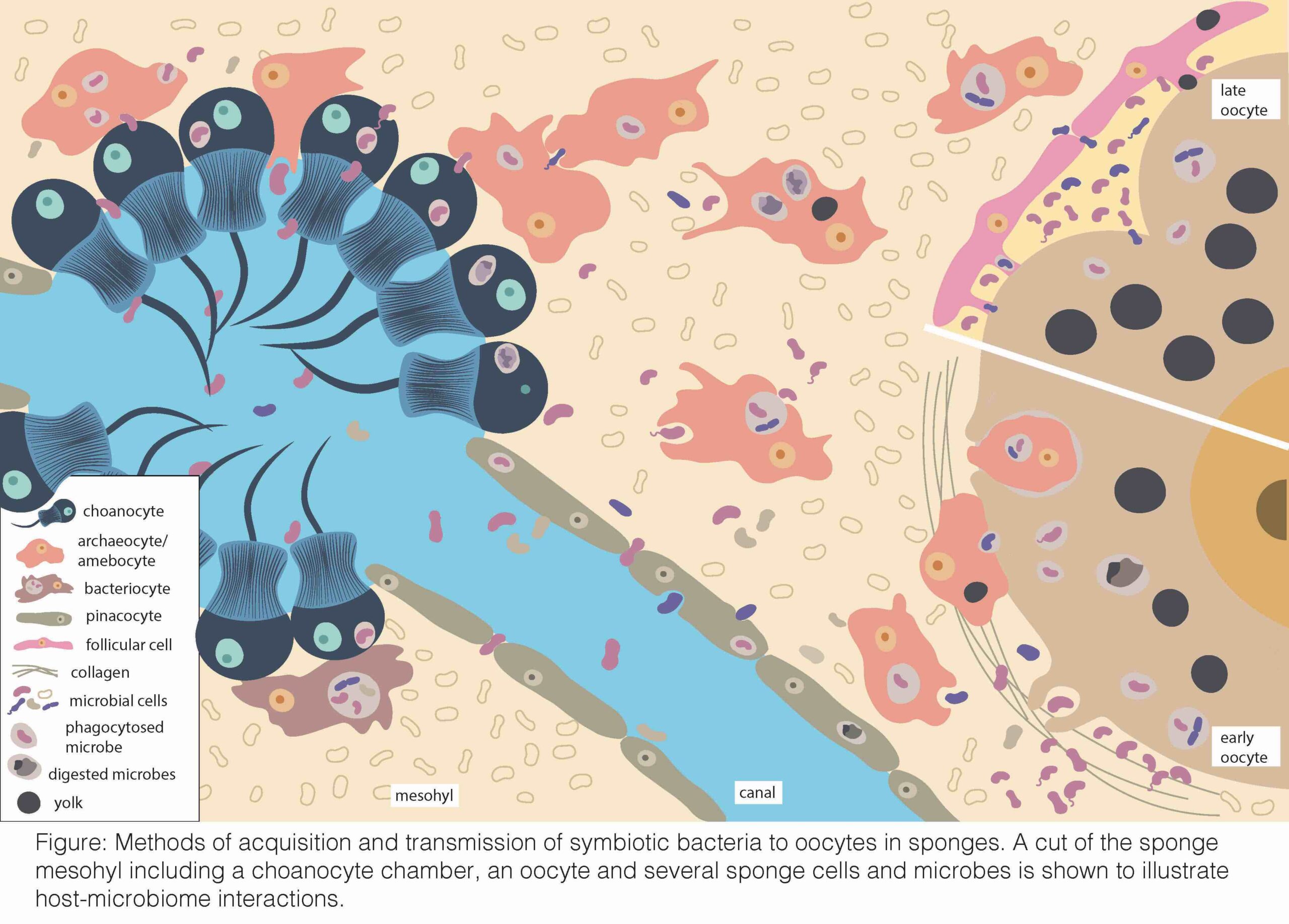
During reproduction, sponges have their highest energetic requirements to produce large quantities of yolk to nourish the oocytes and embryos. However, this becomes challenging because their feeding system, comprised by choanocytes for filtering water and archaeocytes for food distribution, is compromised when these cells transform into the sponge gametes. I aim to understand how the sponge associated microbes support the nutritional status during gametogenesis until the final sponge recruits, and the fidelity in the vertical transmission of these symbionts to offsping. Understanding this host-microbiobe relationship is critical to determine sponge reproductive success, and to predict the effects of ongoing environmental change on the sponge biology, and ultimately the marine ecosystems that they support.
Publications
Recent Publications
Díez-Vives C, Riesgo A. High compositional and functional similarity in the microbiome of deep-sea sponges. ISME Journal 2024, https://doi.org/10.1093/ismejo/wrad030.
Turon M, Ford M, Maldonado M, Sitjà C, Riesgo R. and Díez-Vives C. Microbiome changes through the ontogeny of the marine sponge Crambe crambe. Environmental Microbiome, 2024, 19, 15. https://doi.org/10.1186/s40793-024-00556-7.
Hustus K, Díez-Vives C, Mitsi K, Nutakki J, Kering V, Nguyen IT, Gomes Spencer M, Leys SP, Hill MS, Riesgo A, Hill AL. Algal symbionts of the freshwater sponge Ephydatia muelleri. Symbiosis 2023, 90(3), 259-273.
Díez-Vives C, Koutsouveli V, Conejero M, Riesgo A. Global patterns in symbiont selection and transmission strategies in sponges. Frontiers in Ecology and Evolution 2022, 10, 1015592. https://doi.org/10.3389/fevo.2022.1015592.
Taylor JA, Díez-Vives , Nielsen S., Wemheuer B. and Thomas T. Communality in microbial stress response and differential metabolic interactions revealed by time-series analysis of a sponge holobiont. Environmental Microbiology 2022, 24(5), 2299-2314.. https://doi.org/10.1111/1462-2920.15962.
Group Members
Group Leader
Cristina Díez Vives
Staff scientist
Shaun Nielsen


