Functional and Comparative Multi-omics
RESEARCH GROUPS

David de Juan
Group Leader
Research Summary
For over 500 million years, evolution has shaped animal species, resulting in a vast array of unique traits and molecular configurations. Studying these configurations helps us better understand how different species respond to common survival challenges, such as infectious diseases, genetic disorders, aging, and extreme environmental conditions. Our team integrates cutting-edge “omics” technologies with computational methods to explore evolutionary changes in different animal lineages, focusing on genomic, epigenomic, transcriptomic, and proteomic layers. By building this molecular framework, we aim to guide the development of better strategies for promoting human, animal, and biodiversity health.
Research Lines
Cancer Evolution and Peto’s Paradox in Primates. Cancer is primarily caused by specific mutations that accumulate in somatic cells over a lifetime. As individuals live longer or have more cells, their risk of developing cancer increases. Surprisingly, inter-species differences in longevity and size do not directly influence cancer prevalence—this phenomenon is known as Peto’s paradox. The mechanisms behind Peto’s paradox remain unclear, but the DNA damage response (DDR) plays a crucial role in controlling the accumulation of somatic mutations, and changes in DDR are widespread in species with low cancer prevalence. To investigate its role in Peto’s paradox, we study DDR across different cell types in humans and a panel of nonhuman primates with varying sizes and lifespans. We assess inter-species differences in cell-type-specific responses to mutagens using a combination of computational approaches, cell health assays, and genomic, epigenomic, and transcriptomic profiling. We aim to identify mutations and regulatory changes that control DNA damage and cancer prevalence in species closely related to humans, paving the way for new cancer and aging therapies.
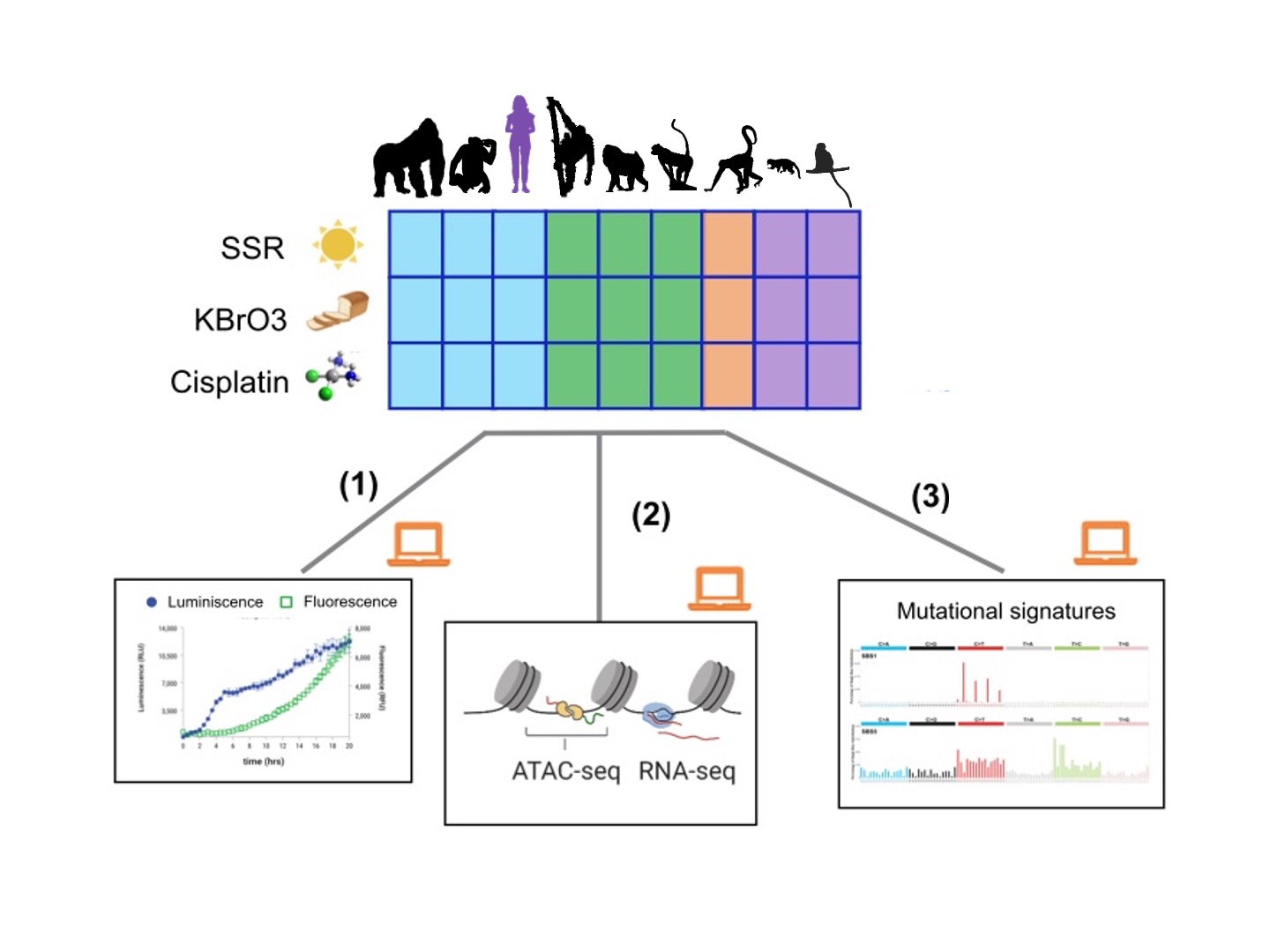
Genetic Determinants of Primate and Mammal Phenome Evolution. The phenome represents the complete set of healthy traits in a species, whether at the level of individuals, tissues, cells, or molecules. These traits arise from molecular processes encoded in the genome and regulated through various mechanisms. Over time, evolution selects phenomes that enhance survival, leaving behind genetic traces in present-day genomes. In collaboration with the Evolutionary Genomics Lab at the Institute of Evolutionary Biology (IBE-CSIC), we conduct comparative studies across species to uncover genetic differences associated with phenotypic variations that have evolved over time. We gather data on hundreds of physical, ecological, and life-history traits from primates and other mammals with high-quality genome sequences. Using advanced computational tools, such as phylogenetic comparative methods, we identify genomic changes that may have influenced key traits in species closely related to humans. These candidate changes are further validated in the lab to confirm their role in shaping observed traits. Our goal is to leverage evolutionary insights to better understand healthy mammalian and primate genomes and phenomes. This knowledge can help inform new treatments for conditions like cancer, aging, neurodegenerative diseases, and rare disorders.
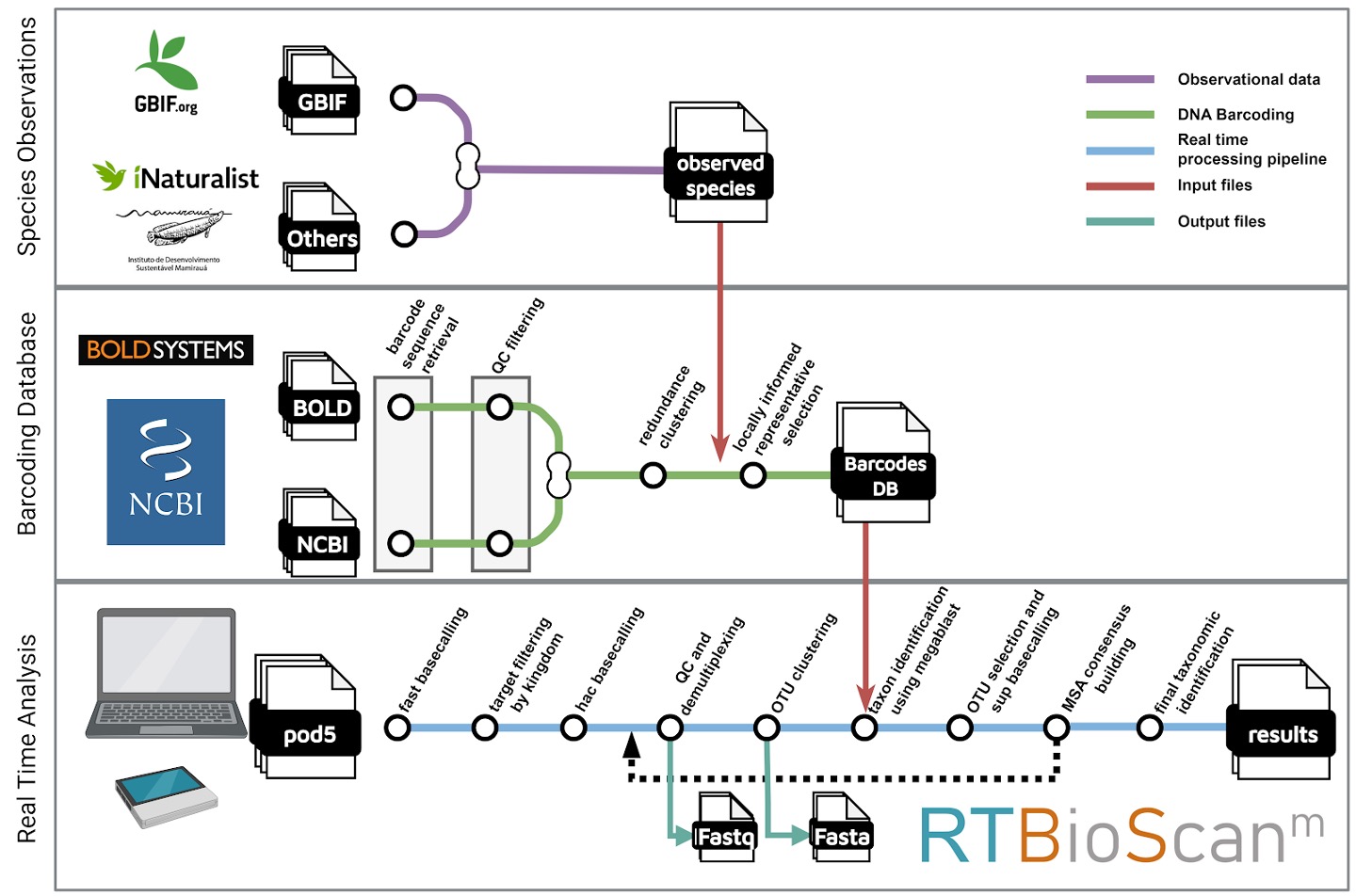
Monitoring Rainforest Biodiversity. Protecting ecosystems that are vital for stabilizing the global climate and preserving biodiversity is more urgent than ever. Rainforests, which play a key role in the planet’s carbon dioxide-oxygen cycle, are home to over 50% of the world’s biodiversity. However, the rapid loss of these still poorly understood tropical forests is pushing many species toward extinction. Our efforts to protect these ecosystems are limited by the lack of effective tools to monitor wildlife at the scale and speed required to prevent further degradation. We are collaborating with the Providence+ team (https://providenceplus.upc.edu/) in the context of the XPRIZE Rainforest challenge (https://www.xprize.org/prizes/rainforest) to develop a scalable solution for monitoring rainforest biodiversity using in situ environmental DNA metabarcoding. In partnership with the Comparative Genomics Lab at the Institute of Evolutionary Biology (IBE-CSIC), the Laboratory of Applied Bioacoustics at the Technical University of Catalonia (UPC), and the Instituto de Desenvolvimento Sustentável Mamirauá (Brazil), we are creating a computational and experimental framework capable of rapidly profiling species in monitored areas through autonomous sampling. This system integrates other biomonitoring technologies used by Providence+, such as bioacoustics and satellite image analysis, to assess the ecological status of these regions. Our framework aims to improve our understanding of rainforest ecological dynamics and empower local communities and governments to manage their natural resources more effectively, supporting conservation and sustainability efforts.
Publications
Group Members
Group Leader
David Juan Sopeña
Funding




