Virus-host Interactions in Hepatitis B Virus Infection
RESEARCH GROUPS

Urtzi Garaigorta
Group Leader
Research Summary
Our laboratory is interested in understanding virus host interactions that regulate the outcome and pathogenesis of virus infections. Our main objective is to identify vulnerabilities that could be exploited to develop new strategies to treat hepatitis B virus infections, that are responsible of acute and chronic hepatitis, cirrhosis and hepatocellular carcinoma. In the last years, we have extended our interest to the development of antiviral treatments to other human viral infections.
Research Lines
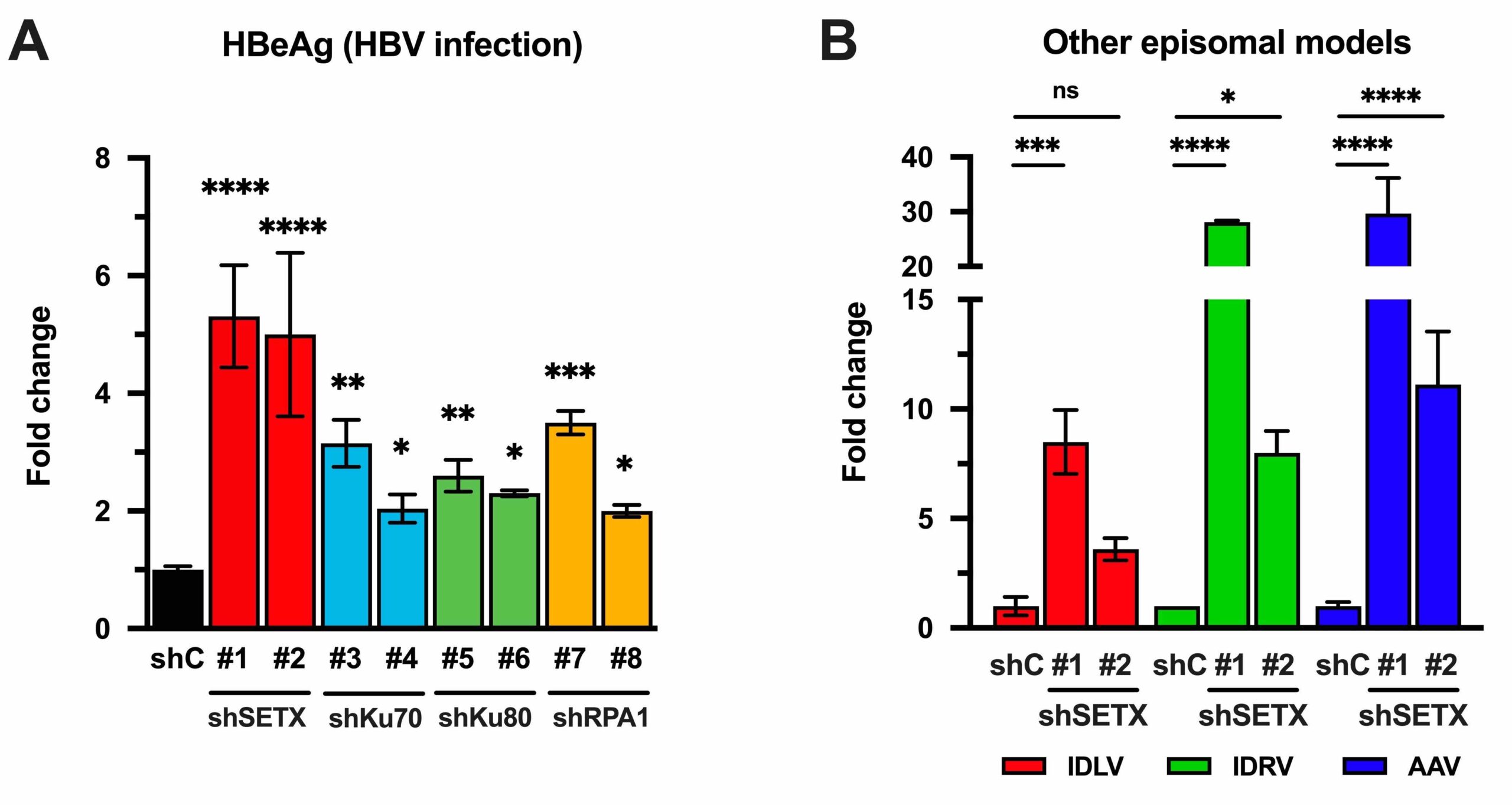
Our research is focused on understanding cellular and molecular mechanisms that regulate hepatitis B virus (HBV) infection, including its pathological consequences. We use cell culture models of HBV infection to study the cellular requirements for the infection as well as to identify host factors that positively or negatively regulate the infection. In the last years, we have identified several cellular proteins (i.e.: Senataxin, Ku70 and Ku80…) that control HBV gene expression by restricting initial steps that are crucial for the establishment of infection.
Effect of topoisomerase inhibitors camptothecin (CPT) and etoposide (Eto) on reporter gene expression (A) and viral DNA integration (B) in hepatic cells transduced with lentiviral vectors that are either competent (ICLV) or deficient (IDLV) in vector integration.
One of our main lines of research is focused on understanding the mechanism(s) by which the identified cellular factors regulate HBV episomal DNA-derived gene expression. To do so, we combine gene silencing and overexpression strategies with the molecular characterization of viral and cellular gene expression analysis, focusing on transcriptomics approaches and chromatin immunoprecipitation analysis. The results from these studies point to a selective regulation of episomal DNA expression, which opens a venue for development of novel therapeutic strategies.
More recently we have extended these observations to other episomal DNA viral models including adeno-associated viral vectors that are being used in gene therapy. Our main objective here is to increase our knowledge of how episomal DNA-derived expression is regulated in order to develop new adjuvants for gene therapy. To complement these studies, we have developed a dual fluorescence reporter screening assay to simultaneously quantify the expression from episomal and chromosomal DNA templates, which in turn allows the identification of small molecules that selectively regulate episomal DNA-derived gene expression.
Finally, our group is also involved in the Antiviral Identification and Characterization Platform of the CNB, which pursues the development of novel antivirals against viruses with biomedical interest including: hepatic viruses, influenza viruses, viruses transmitted by mosquitos, coronaviruses…
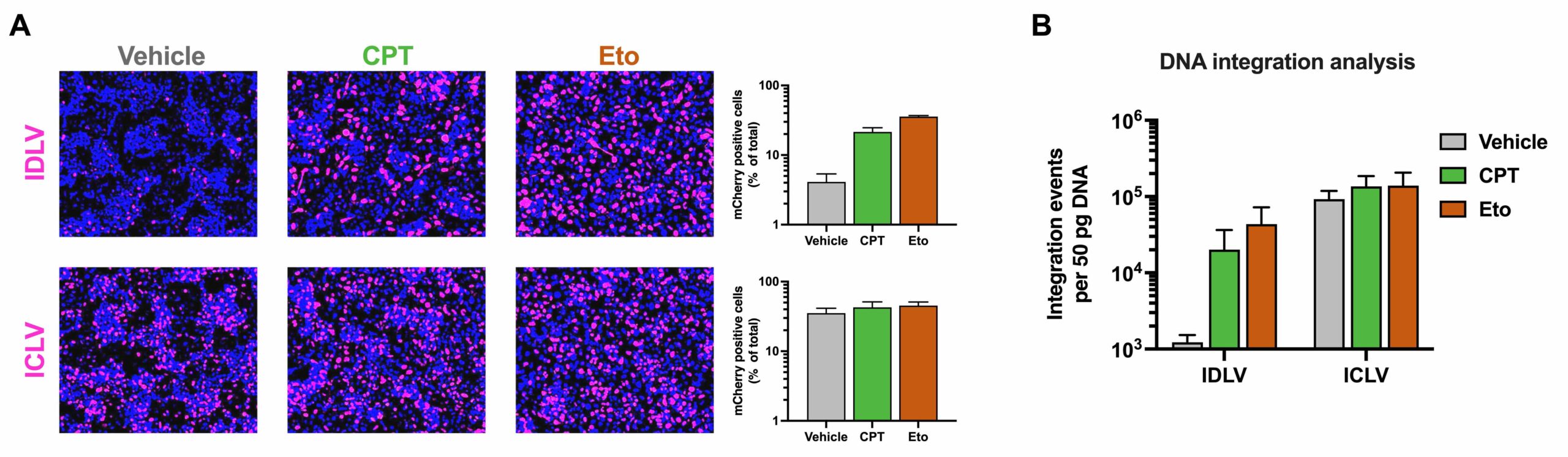
Effect of topoisomerase inhibitors camptothecin (CPT) and etoposide (Eto) on reporter gene expression (A) and viral DNA integration (B) in hepatic cells transduced with lentiviral vectors that are either competent (ICLV) or deficient (IDLV) in vector integration.
Publications
Group Members
Group Leader
Urtzi Garaigorta
Lab assistant
Jennifer Moya Vaquero
Postdoctoral Researcher
Beatriz Chamorro Gorines
PhD candidates
Enara San Sebastian Casas
Adrián Coto Ramos
Rubén Gutierrez Molina

Funding
News
Real-space heterogeneous reconstruction, refinement, and disentanglement of CryoEM conformational states with HetSIREN
Nat Commun. 2025 Apr 22;16(1):3751. Herreros D, Mata CP, Noddings C, Irene D, Krieger J, Agard DA, Tsai MD, Sorzano COS, Carazo JM. Abstract Single-particle analysis by Cryo-electron microscopy (CryoEM) provides direct access to the conformations of...
Ceftazidime-avibactam use selects multidrug-resistance and prevents designing collateral sensitivity-based therapies against Pseudomonas aeruginosa
Nat Commun. 2025 Apr 9;16(1):3323. Hernando-Amado S, Gomis-Font MA, Valverde JR, Oliver A, Martínez JL Abstract Ceftazidime-avibactam is a β-lactam/β-lactamase inhibitor combination restricted for the treatment of multidrug-resistant infections of Pseudomonas...
El Centro Nacional de Biotecnología estrena nueva imagen para reflejar una etapa de renovación y evolución
El nuevo logotipo está compuesto por una espiral que representa una hélice de ADN vista desde arriba, inspirada en la emblemática “Fotografía 51” de Rosalind Franklin, clave para desentrañar su estructura La presentación de esta imagen coincide con el Día del ADN, una...





