Bacteria-based Cancer Immunotherapies
RESEARCH GROUPS

Esteban Veiga
Group Leader
Research Summary
We aim to shape the next generation of cancer immunotherapies. To achieve this goal and surpass current therapies, we are leveraging the abilities of certain bacteria to enhance immune responses.
Research Lines
We are focus in generating novel immunotherapies using the ability of bacteria to modify the immune responses. We have discovered that CD4+ T cells contributes to the early immune response capturing bacteria by transfagocytosis. Surprisingly, the transfagocytic CD4+ T cells destroy bacteria and become hyperinflammatory (Cruz-Adalia et al. 2014). Moreover, we have discovered that bacteria exposure “instructs” conventional CD4+ T cells. Instructed CD4+ T cells (bacT), contrary to the role separation dogma in immunology, became potent antigen-presenting cells able to cross-present antigens from captured bacteria, activating naïve CD8+ T cells that became effective cytotoxic cells (Cruz-Adalia et al. 2017). Similarly, B cells can be also instructed by bacteria (García-Ferreras et al 2023)
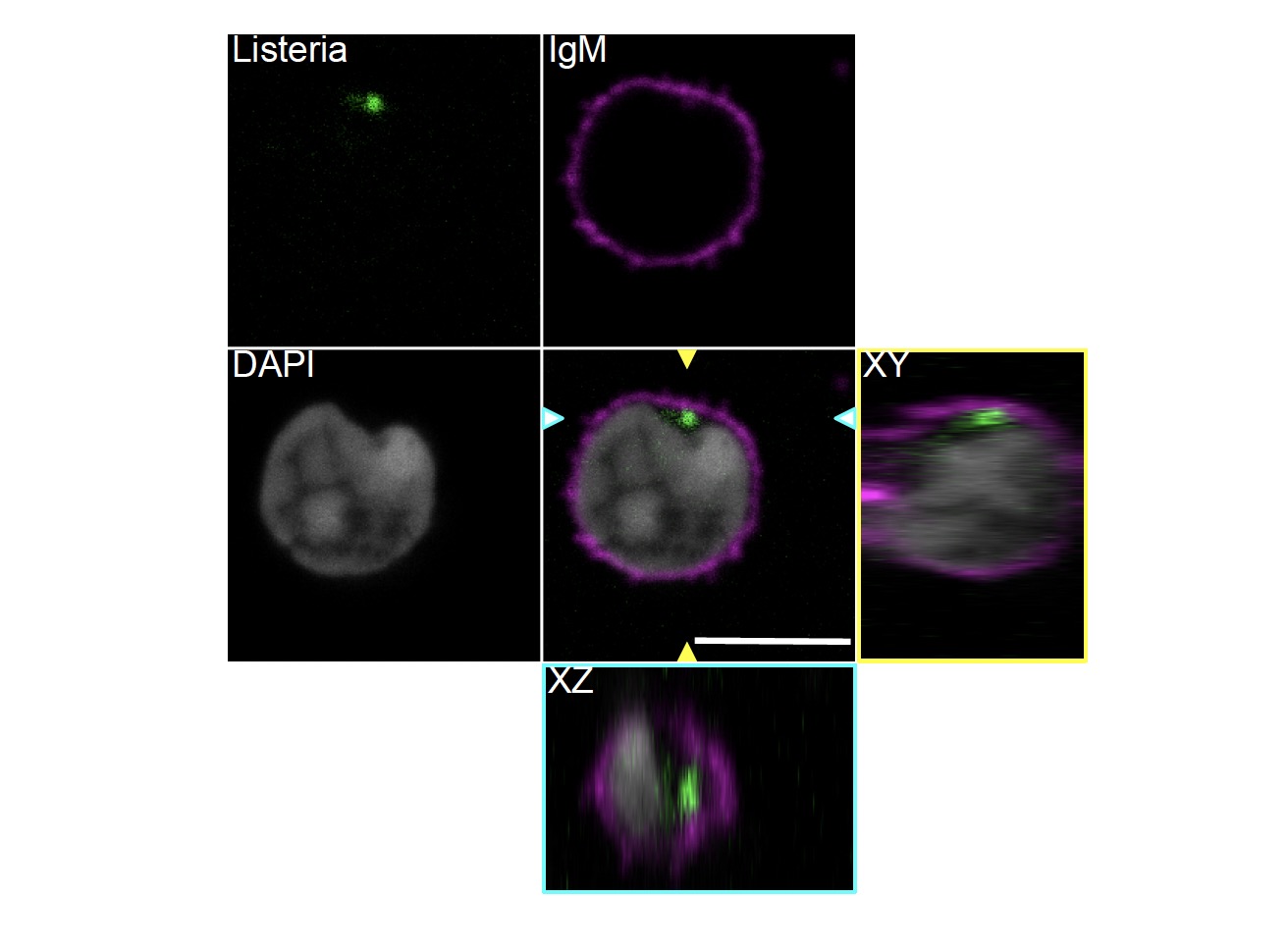
Confocal images of B cells from WT mice that have captured L. monocytogenes (green), after 4 hours of infected DC/B cell contacts. B cell surface was detected using an anti IgM antibody (magenta) the B cell nucleus was stained with DAPI (cyan in B and grey in C). The orthogonal views show intracellular bacteria. Scale bar represents 5 mm. Arrowheads indicate the XZ and YZ axes. From García-Ferreras R., et al 2023.
Proof-of-concept experiments show that mice treated with bacT cells that have captured/killed bacteria expressing tumor antigen were protected against tumor development (Cruz-Adalia et al. 2017). These discoveries challenged the dogma of adaptive/innate immunity role separation and guided our current research to find novel tumor immunotherapies.
In collaboration with Drs Carlos O. S. Sorzano (CNB) and Arrate Muñoz Barrutia (Univ Carlos iii) we developed a novel platform to identify tumor neoantigens from NGS data based in machine learning (Wert et al 2021; Méndez-Pérez et al 2024)
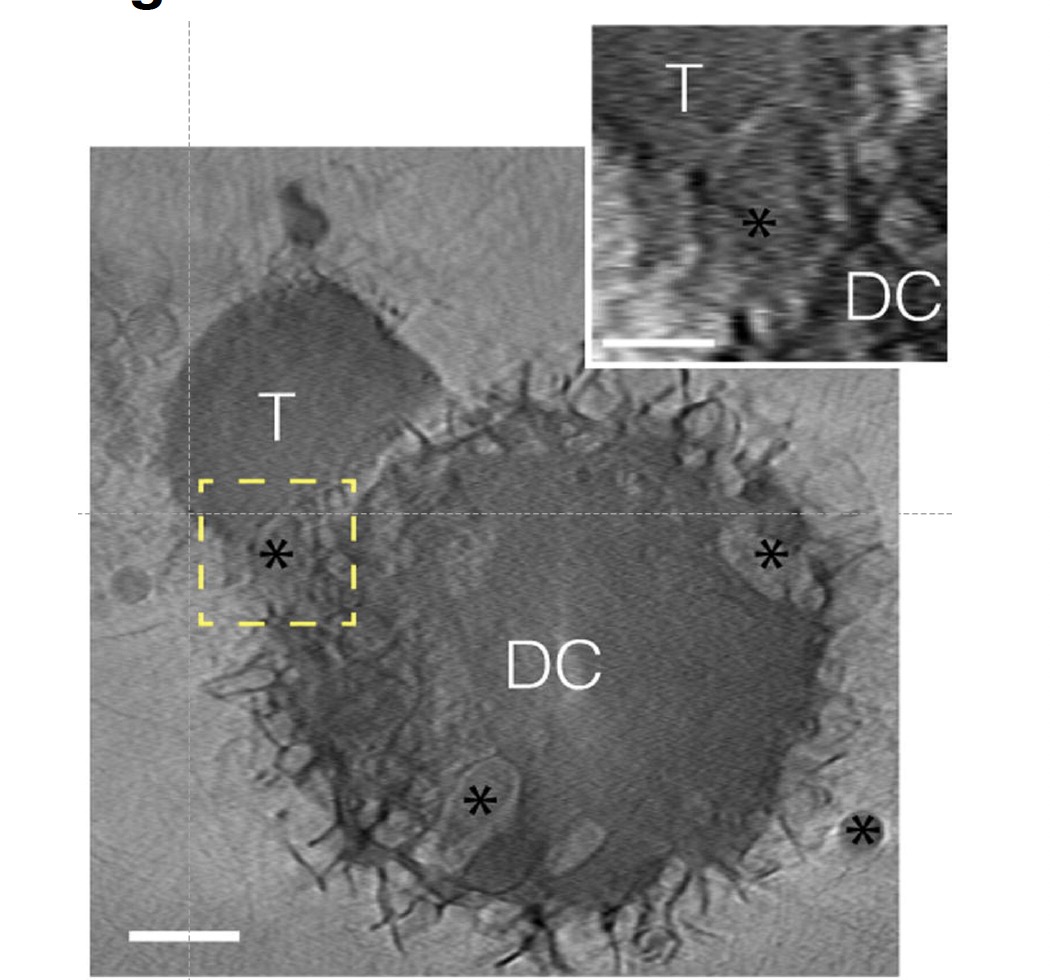
Virtual slices of a cryo soft X-ray tomogram acquired at BESSY-II synchrotron (Berlin, Germany) from a T cell (T) interacting with an S. enterica-infected DC. The asterisks mark the position of Salmonella. The indicated bacteria are inside DC close to the IS (left) and outside DC and being engulfed by the T cell (right). Inset shows a detailed position of bacteria. Scale bars represent 2 micron and 1 micron in the insets. From Cruz-Adalia et al 2014.
Publications
Group Members
Group Leader
Esteban Veiga
PhD candidates
Daniel Gómez Garrido
Inés de Cáceres Renovell

Funding
News
Las Sinergias Severo Ochoa financiarán 8 proyectos colaborativos de grupos del CNB
La convocatoria interna Sinergias Severo Ochoa del Centro Nacional de Biotecnología (CNB-CSIC) financiará ocho proyectos colaborativos de grupos de investigación del centro, impulsando el desarrollo de programas transversales de alta calidad científica. Entre las...





