Chemokine Receptors as targets for cancer and inflammation therapy
RESEARCH GROUPS

Mario Mellado
Group Leader
Research Summary
Through their interactions with G protein-coupled receptors, the chemokines have a key role in a broad array of biological responses that include cell polarisation and movement, immune and inflammatory responses, haematopoiesis, tumour rejection and prevention of HIV-1 infection.
In WHIM syndrome, mutations in the CXCR4 receptor affect the formation of clusters in the cell membrane necessary to respond to the “call” of the chemokine CXCL12. This results in changes in the actin cytoskeleton, involving the β1-arrestin protein, the cell loses its sense of orientation and no longer follows the direction of the chemoattractant gradients. WT cells (left video) vs Mutant cells (right video). PNAS 2022
Research Lines
Cell migration involves myriad signaling proteins and receptors that act coordinately to activate intracellular pathways and promote polarized cell states and directional migration. To migrate directionally in response to external stimuli, the internal machinery of cells needs to be spatially organized, which involves the integration of biochemical and mechanical factors to generate force in a specific direction to move the cell forward. Chemokine receptors are membrane-expressed seven-transmembrane receptors linked to G proteins. Through interaction with the corresponding ligands, they induce a wide variety of cellular responses including cell polarization, movement, immune and inflammatory responses, as well as prevention of HIV-1 infection. For achieving their function, chemokine receptors undergo ligand-dependent conformational changes that are influenced by the local cellular microenvironment.
Using quantitative single-molecule spatio-dynamic imaging, our group studies the dynamics of the chemokine receptors at the cell membrane and the factors that influence it. We observed how the maintenance of correct actin dynamics in migrating cells is essential for the formation of large receptor nano-clusters in response to the ligand and its role for a correct sensing of the chemoattractant gradients. We have determined that altered cluster formation, using natural mutant chemokine receptors or altering the lipid composition of the cell membrane, results in the inability of these cells to correctly sense chemokine gradients. Using in silico strategies and in vitro assays we have also designed some allosteric modulators that block receptor clustering and directed cell migration without altering neither ligand binding nor other signaling events. The evidence indicates that receptor clustering can be exploited as therapeutic target in inflammatory diseases and cancer.
In addition, our group is also involved in studying the effect of distinct mediators in reprograming/training of macrophages to improve resolution of some disease.
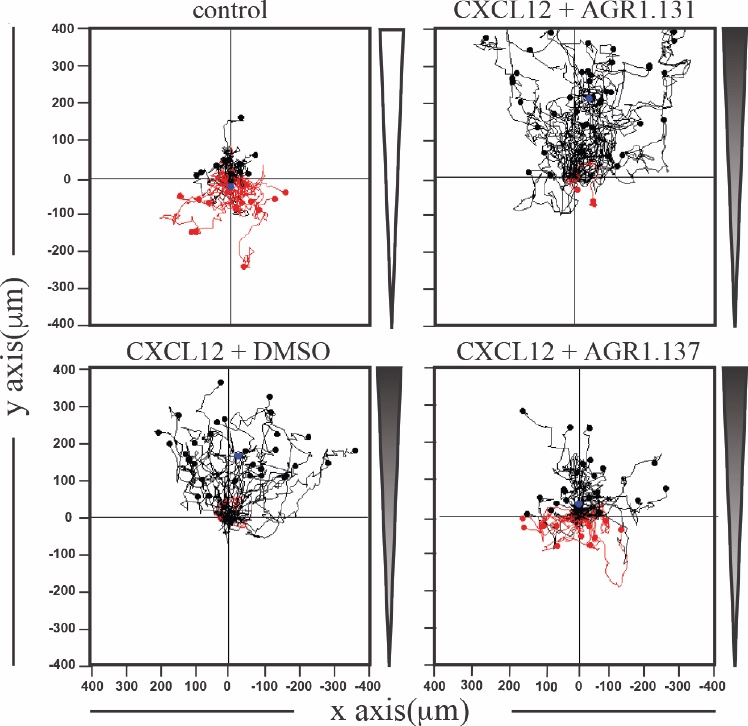
Migration of HeLa cells treated with vehicle (DMSO) or with the selected small compounds (AGR1.131 or AGR1.137) as indicated, on μ-chambers in response to a CXCL12. Figure shows representative spider plots with the trajectories of tracked cells migrating along the gradient (black) or moving in the opposite direction (red).
Using several techniques based in advanced-light and correlative microscopy our group studies:
- How membrane fluidity alters CXCL12-mediated CXCR4 dynamics: effect of cholesterol depletion
- The consequences of HIV-1 binding on the clustering and dynamics of CXCR4 at the cell membrane. Effects on HIV-1 infection
- The influence of ACKR3 on CXCR4 oligomerization and dynamics. ACKR3 is an atypical receptor that does not activate G proteins but oligomerizes with CXCR4
- The role of CCL19 and CCL21 to promote CCR7 nanoclustering and its influence on T cell migration
- Chemokine receptor nanoclustering at the immunological synapse (IS) and its functional relevance in T cell activation
- The effect of the chemokine CCL2 on macrophage metabolism and in the structure of mitochondrias
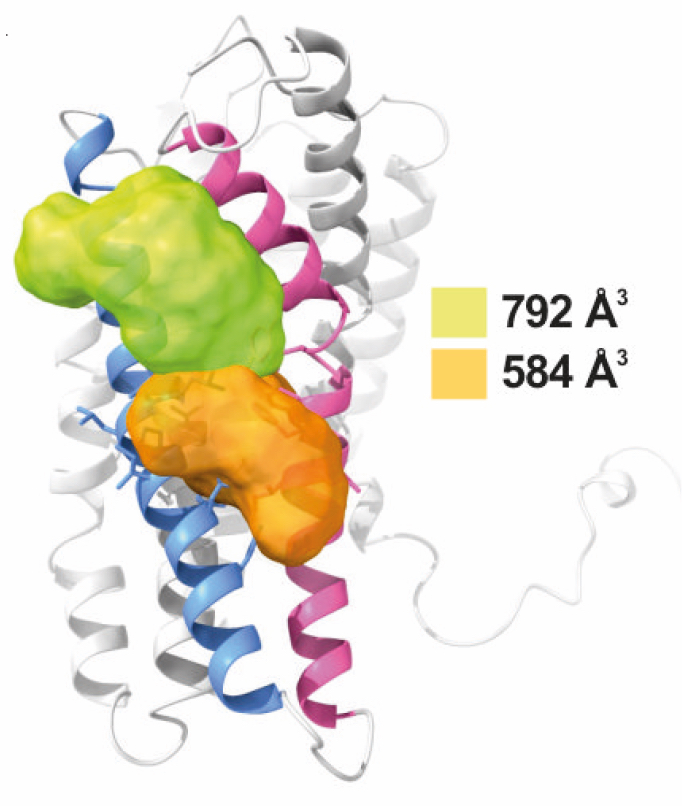
Surface representation of CXCR4 and the clefts identified between transmembrane (TM) region V and VI. The protein structure is shown in gray, with TMV and TMVI colored in blue and pink respectively. The small compounds AGR1.135 and AGR1.137 binding, were identified by SurfNet software, are shown in orange and green respectively. Volumes for each cavity are measured (Å3)
Publications
Group Members
Group Leader
Mario Mellado
Project Leader
José Miguel Rodríguez Frade
Staff scientists
Eva Mª García Cuesta
Blanca Soler Palacios
PhD candidates
Noelia Santander
Jesús del Amo

Funding
Our research is funded by national and international institutions as indicated below. For more details, please check the general Funding Section at the CNB website.
News
Expresión de interés en doctorado en el grupo “receptores de Quimioquinas”
IP: Mario Mellado, Research group Buscamos candidatos/as entusiastas y altamente motivados para solicitar una beca predoctoral en las convocatorias que se encuentran abiertas actualmente y realizar una tesis doctoral en nuestro laboratorio.Características del grupo:En...
Identifican un compuesto capaz de modular la migración celular en procesos autoinmunes y tumorales
19 de Septiembre 2024 El compuesto hallado por un equipo del CSIC regula la capacidad de inducir la migración celular en el receptor de las quimioquinas, unas proteínas implicadas en procesos inflamatorios, autoinmunes o de metástasis tumoral Este trabajo abre la...
FPI contract associated with the NANHOMIS project (PID2023-146301OB-I00)
Título del proyecto: “Role of Chemokine receptor nanoclustering in T cell homing to lymph nodes and Immunological synapses formation (NANHOMIS)”. IP: Mario Mellado, Research group Reference: PID2023-146301OB-I00 We are seeking enthusiastic and highly motivated PhD...



