Transcription-Replication Conflicts in Eukaryotes
RESEARCH FELLOW

Esther Ortega
Ramón y Cajal Fellow
Research Summary
Our group aims to understand the molecular mechanisms that maintain eukaryotic genome stability during DNA replication, using a combination of biochemical, biophysical, cellular and structural techniques.
Research Lines
Our group has been established in 2021 and aims to understand the molecular mechanisms that maintain eukaryotic genome stability during DNA replication, using a combination of biochemical, biophysical, cellular and structural techniques. DNA replication and RNA transcription are two essential processes required for accurate cell function and the propagation of genetic information. As both machineries need to access to the same DNA substrate, efficient coordination between these two processes is essential to maintain the integrity of the genome. However, these machineries can meet in space and time, causing transcription-replication conflicts (TRCs), which are a main cause of genomic instability. In eukaryotic organisms, TRCs interfere with the progression and stability of the replication forks and also trigger the accumulation of dangerous recombinant DNA structures (as R-loops) which slows down or stalls replication forks due to the physical impediments that prevents its advance. Stalled forks are a threat for DNA duplication and genome stability, causing neurodegeneration and cancer. Our group is focused on understanding the role of several factors important in the resolution of TRCs. We aim to elucidate how human Senataxin and PCNA_unloading complexes protects the integrity of the replication machinery and resolve R-loops structures formed during these TRCs.
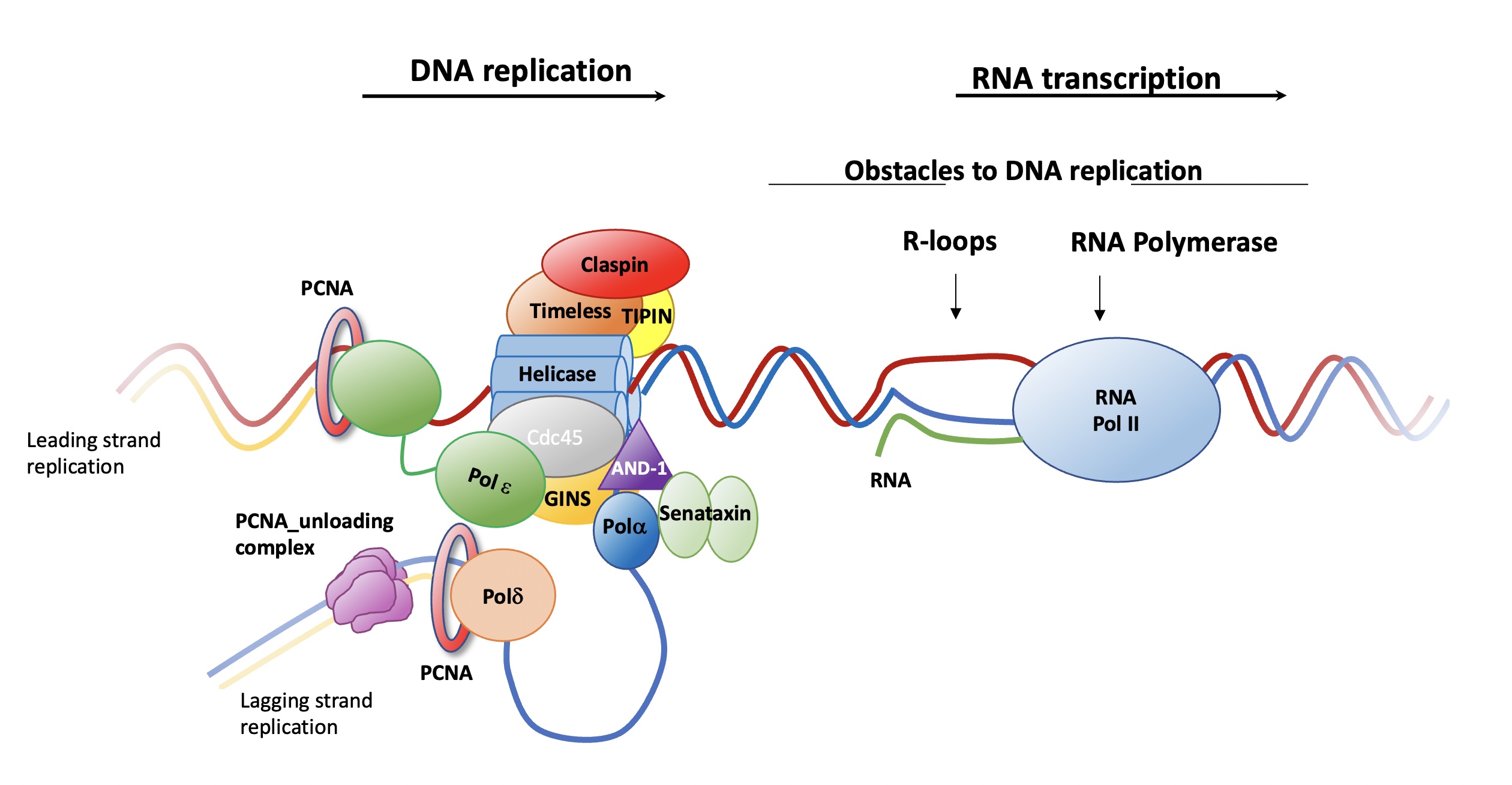
The DNA replication machinery (replisome) is a multi-protein complex which must overcome diverse ‘obstacles’ as R-loops and RNA Polymerase to faithfully replicate DNA. This is done by recruiting other factors, as Senataxin or PCNA-unloading complex. Our group investigates the molecular mechanisms of the function of these proteins.
Publications
Group Members
Group Leader
Esther Ortega
Technician
Eduardo Muñoz
PhD candidate
José Manuel Aguilella
Funding
Our research is funded by national and international institutions as indicated below. For more details, please check the general Funding Section at the CNB website.
News
Cytolytic γδ T-cells and IFNγ-producing CD4-lymphocytes characterise the early response to MTBVAC tuberculosis vaccine
NPJ Vaccines. 2025 Mar 28;10(1):58. Felgueres MJ, Esteso G, García-Jiménez ÁF, Benguría A, Vázquez E, Aguiló N, Puentes E, Dopazo A, Murillo I, Martín C, Rodríguez E, Reyburn HT, Valés-Gómez M Abstract Infection with Mycobacterium tuberculosis (Mtb) can produce...
Replicating community dynamics reveals how initial composition shapes the functional outcomes of bacterial communities
Nat Commun. 2025 Mar 31;16(1):3002. Pascual-García A, Rivett DW, Jones ML, Bell T. Abstract Bacterial communities play key roles in global biogeochemical cycles, industry, agriculture, human health, and animal husbandry. There is therefore great interest in...
Rational Design Assisted by Evolutionary Engineering Allows (De)Construction and Optimization of Complex Phenotypes in Pseudomonas putida KT2440
Microb Biotechnol. 2025 Mar;18(3):e70132. Blázquez B, Nogales J. Abstract Beyond the rational construction of genetic determinants to encode target functions, complex phenotype engineering requires the contextualisation of their expression within the metabolic and...



