Microbial Populations and Comparative Genomics
RESEARCH GROUP
Research Summary
Our research group focuses on the comparative genomics and population genomics of microorganisms to understand their diversity, evolution, and functional potential. By studying microbial populations across different environments, we aim to uncover the genetic basis of microbial adaptability, resilience, and specialization. Comparative genomic approaches allow us to identify key genetic traits that drive these processes, offering valuable insights into microbial ecology and evolution.
In addition to these genomic studies, our group is particularly interested in the interactions between microorganisms and plants. Microbes play crucial roles in plant health and growth, influencing nutrient cycling, disease suppression, and stress tolerance. By exploring how microbial populations interact with plant hosts, we aim to uncover mechanisms that could lead to sustainable agricultural practices and improve plant resilience in diverse ecosystems. Our research brings together the power of genomics with the complexity of plant-microbe interactions to address key challenges in both microbial ecology and plant science.
Research Lines
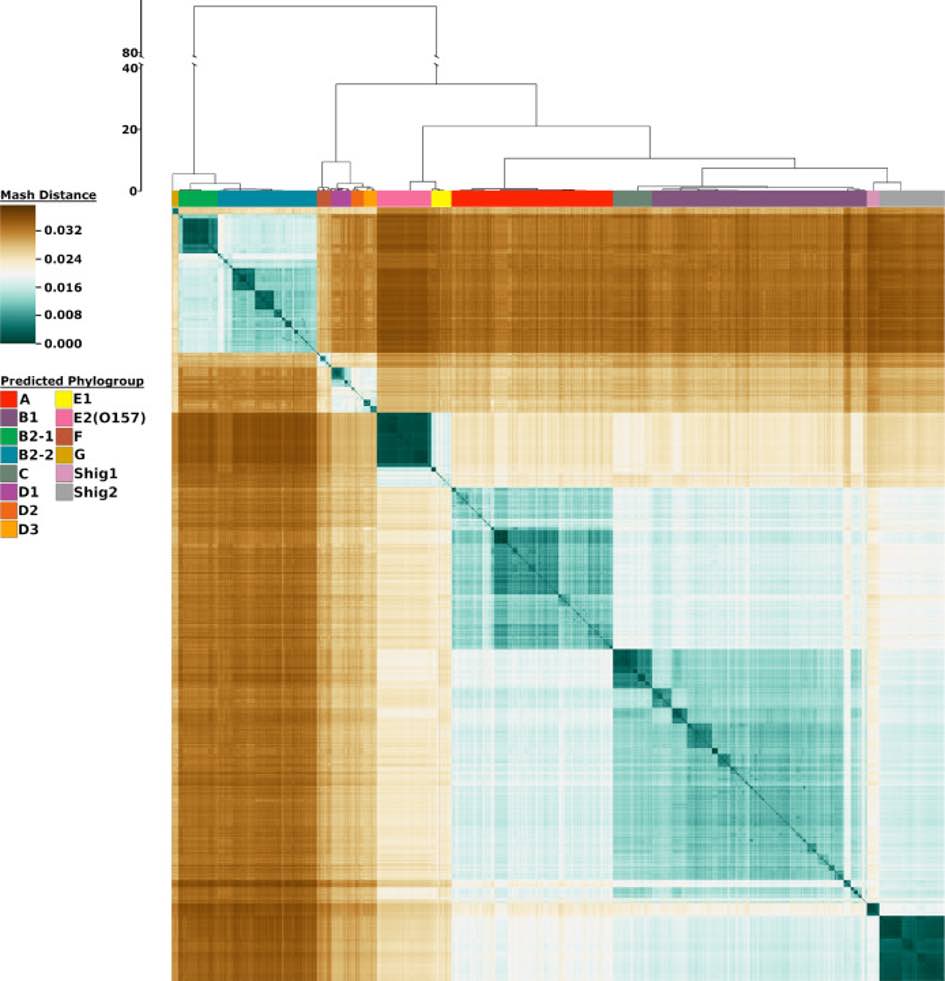
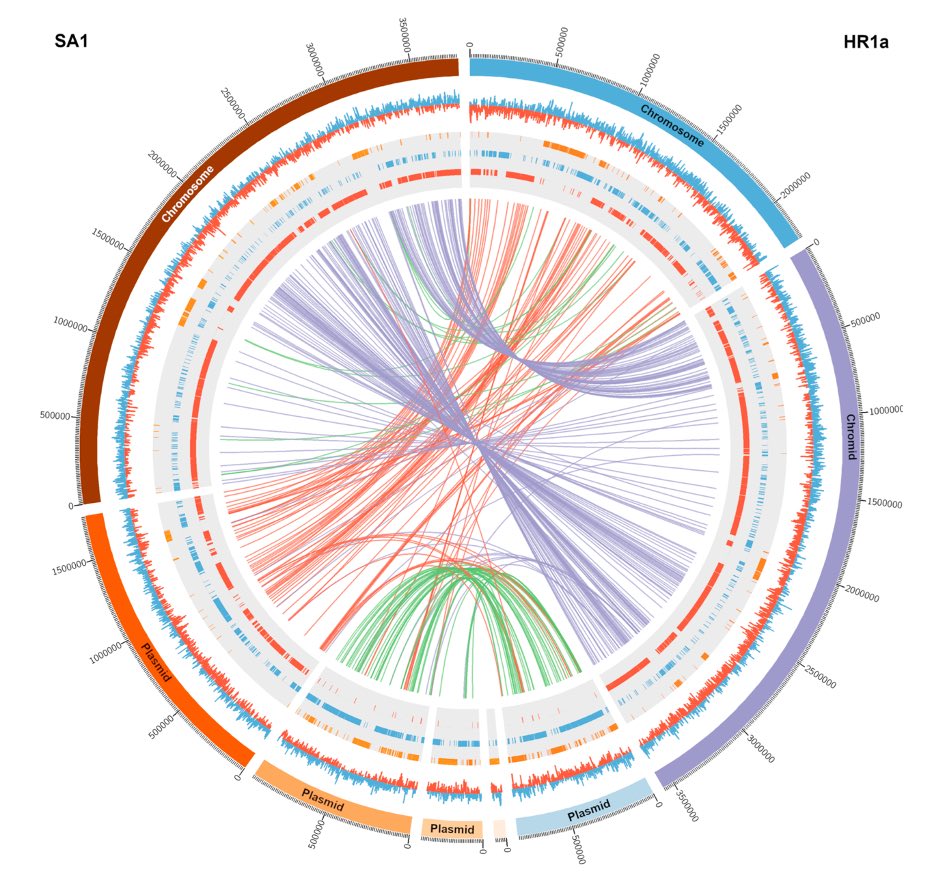
Moreover, by analyzing the population structure of microbial species, it is possible to identify patterns of genetic variation across different ecological niches, helping to elucidate the evolutionary processes shaping microbial communities. As high-performance computational tools and sequencing technologies continue to evolve, comparative genomics has become an indispensable approach for exploring microbial populations and their ecological dynamics, contributing to fields such as microbial epidemiology, evolution, and synthetic biology.
Scientific Publications
Group Members
Zulema Udaondo
Funding

News
Real-space heterogeneous reconstruction, refinement, and disentanglement of CryoEM conformational states with HetSIREN
Nat Commun. 2025 Apr 22;16(1):3751. Herreros D, Mata CP, Noddings C, Irene D, Krieger J, Agard DA, Tsai MD, Sorzano COS, Carazo JM. Abstract Single-particle analysis by Cryo-electron microscopy (CryoEM) provides direct access to the conformations of...
Ceftazidime-avibactam use selects multidrug-resistance and prevents designing collateral sensitivity-based therapies against Pseudomonas aeruginosa
Nat Commun. 2025 Apr 9;16(1):3323. Hernando-Amado S, Gomis-Font MA, Valverde JR, Oliver A, Martínez JL Abstract Ceftazidime-avibactam is a β-lactam/β-lactamase inhibitor combination restricted for the treatment of multidrug-resistant infections of Pseudomonas...
El Centro Nacional de Biotecnología estrena nueva imagen para reflejar una etapa de renovación y evolución
El nuevo logotipo está compuesto por una espiral que representa una hélice de ADN vista desde arriba, inspirada en la emblemática “Fotografía 51” de Rosalind Franklin, clave para desentrañar su estructura La presentación de esta imagen coincide con el Día del ADN, una...



