Genetic Stability
RESEARCH GROUPS

Juan Carlos Alonso
Group Leader

Silvia Ayora
Group Leader
Research Summary
Our goal is to study proteins that control genetic stability using Bacillus subtilis as a model.
Research Lines
Transcription, replication and repair occur concurrently on shared DNA template in actively growing bacterial cells, making their coordination essential. When the replisome or the RNA polymerase elongation complex encounters DNA damage, replication and transcription stall, leading to traffic conflicts. These encounters between the two machineries may result in disruption of replication and genome instability. Then recombination and repair proteins become essential to maintain genome stability.
Our research focuses on the study of the proteins that secure genomic stability using Bacillus subtilis (a representative bacteria of the Firmicutes phylum) as a model. We analyze how DNA helicases and nucleases that contribute to resolve transcription-replication conflicts may act on stalled replication forks.
In addition, we have analyzed a three-component ParABS partition system, present in the low-copy number plasmid pSM19035. Our results reveal how the ParA and ParB proteins may ensure the stable inheritance of many bacterial chromosomes and low-copy-number plasmids.
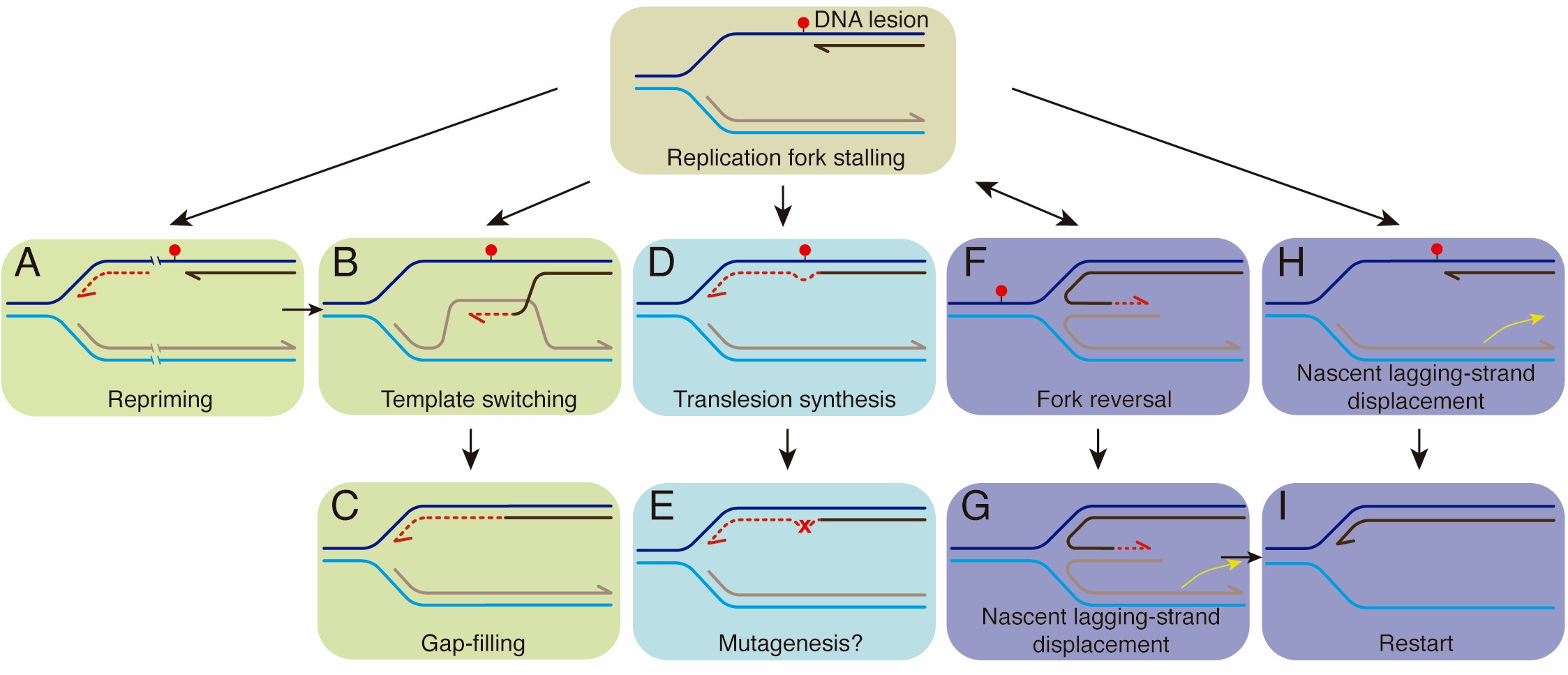
Potential replication stress response mechanisms after DNA replication stalling. A replicative DNA polymerase cannot accommodate a damaged template base (represented by a red dot) and transiently stalls. Replication may proceed via error-free (template switching, fork remodeling) or error-prone pathways to allow DNA replication to resume (A-E) or restart (F-I).
Publications
Group Members
Group Leaders
Juan Carlos Alonso
Silvia Ayora
Staff scientist
Begoña Carrasco
Rubén Torres
Lab assistant
María López



