Environmental Synthetic Biology
RESEARCH GROUPS

Víctor de Lorenzo
Group Leader
Research Summary
The longstanding mission of our team is the genetic programming of bacterial agents for biosensing, large-scale remediation and valorisation of chemical waste that is otherwise dumped into the environment by urban and industrial activities. To this end, we have adopted the conceptual frame and the experimental approaches of contemporary synthetic biology. Our biological platform of choice is the soil bacterium Pseudomonas putida, which combines the ease of genetic and genomic editing that is typical of Escherichia coli with the safety, robustness and metabolic capabilities required in whole-cell catalysts for applications in harsh biotechnological settings.
Research Lines
[i] Development of P. putida as a reliable chassis for implantation of genetic and metabolic circuits. This involves a profound editing of the extant genome of this microorganism for enhancing desirable properties and eliminating drawbacks. Also, the exploitation of surface-display systems for designing complex catalytic properties altogether separated from the cell metabolism and even the design of artificial communities by means of ectopic adhesins.
[ii] Genetic tools for deep refactoring of metabolic and physical properties of P. putida. The list of new assets that we are developing includes a large collection of standardized plasmid and transposon vectors as well as dedicated reporter systems for parameterization of the gene expression flow and for switching entire metabolic regimes. The central resource to this end is the so-called SEVA platform (http://seva-plasmids.com)
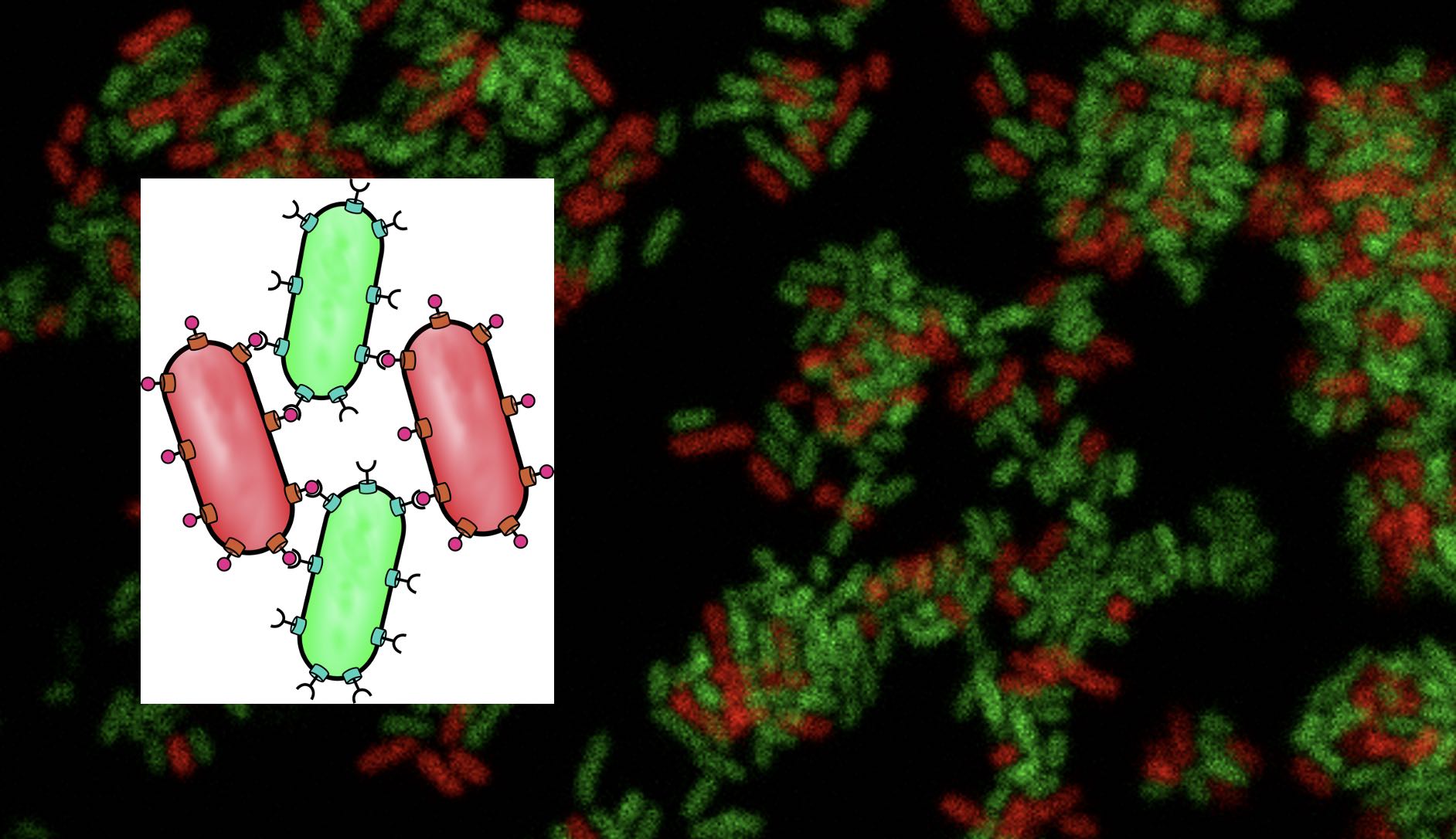
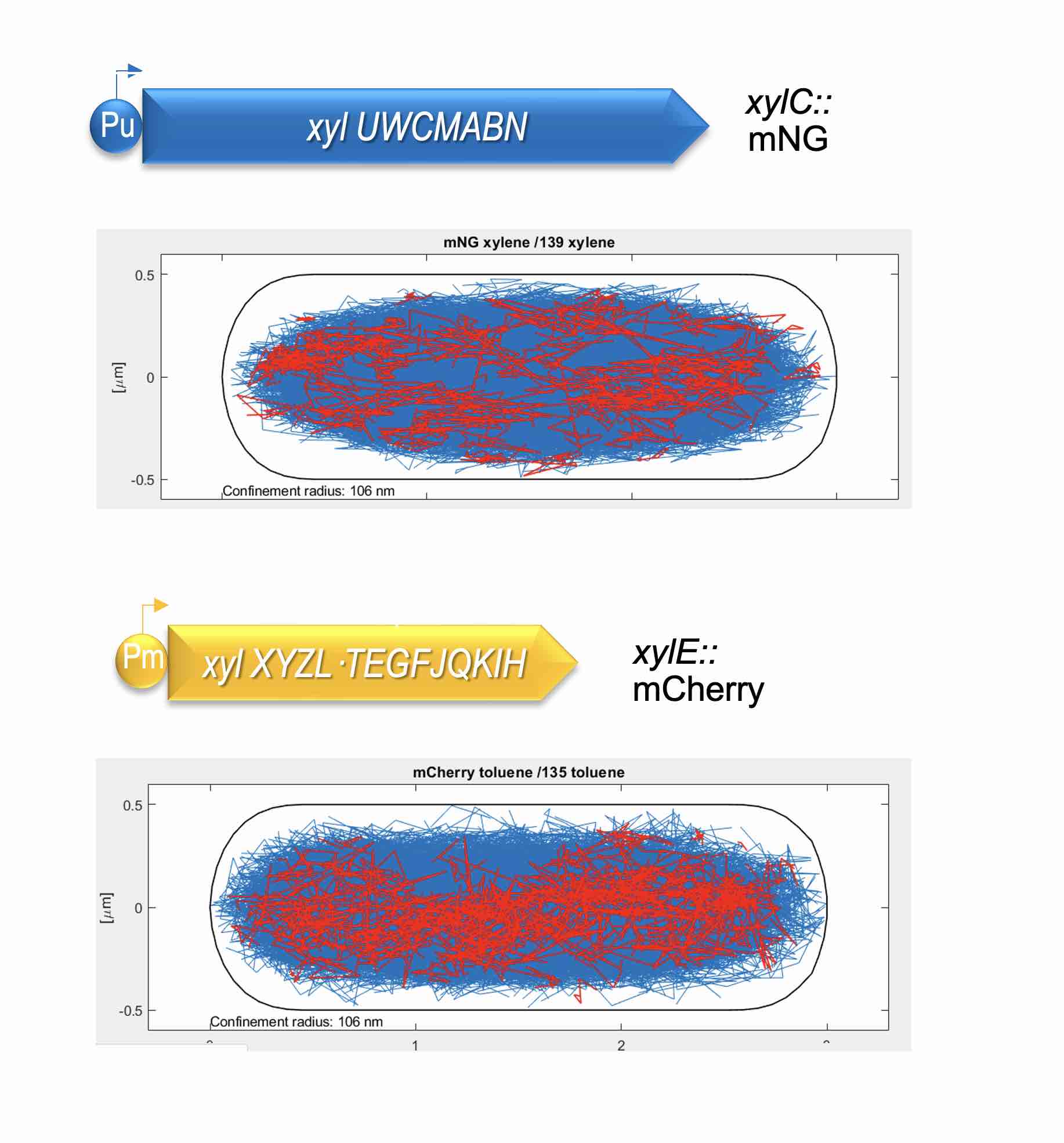
[iii] The TOL system borne by plasmid pWW0 as a natural example of well-nested metabolic circuit implantation. The two operons for toluene and m-xylene biodegradation encoded in pWW0 offer a case of expansion of the metabolic repertoire of environmental bacteria through acquisition of new genes. Furthermore, intracellular localization of the TOL-encoded proteins exposes the physical dynamics of a complex metabolic process.
[iv] Deep metabolic engineering of P. putida. Currents efforts attempt to develop strains that can be entirely programmed to behave as whole-cell catalysts upon exposure and computation of both external and internal cues. This endeavour combines direct rational engineering with fine-tuning of gene expression by means of site-specific diversification of genomic sequences of choice. Multi-objective optimimization is typically brought about by adaptive evolution.
Publications
Group Members
Group Leader
Víctor de Lorenzo
Associate Researcher
Sebastian Wenk
Project leader
Esteban Martínez
David Rodríguez
Staff scientists
Belén Calles
Tomás Aparicio
Elena Algar
Lab manager
Inés Merino
Postdoctoral scientist
Guillermo Gómez
PhD candidate
Francisco Moreno
Lab assistants
Sofía Fraile
Irene del Olmo

Funding





