Dynamics of RNA Viruses in Infected Patients. New Antiviral Designs
RESEARCH GROUP

Celia Perales Viejo
Group Leader
Research Summary
Virus variability is one of the main obstacles for the effective prevention and treatment of viral diseases. The main objective of our laboratory is to understand viral dynamics of RNA viruses, based on deep sequencing data of in vivo and ex vivo viral populations. We aim at designing new antiviral strategies capable of counteracting virus adaptability which is responsible of treatment and vaccination failures. We are currently analyzing SARS-CoV-2 from nasopharyngeal swabs of the several pandemic waves, in the search of synergistic antiviral combinations to suppress viral replication, including the use of nucleoside analogues that act as lethal mutagens.
Research Lines
Viral quasispecies refers to a population structure that consists of extremely large numbers of variant genomes, termed mutant spectra, mutant swarms or mutant clouds that are subjected to genetic variation, competition and selection. Due to the high mutation rates, mutants arise continually, and they change in relative frequency as viral replication proceeds. This introduces an indetermination regarding the precise composition of viral populations. Genomes present at low frequency in a mutant spectrum fuel virus adaptability, and may also influence the behavior of the population ensemble. Selective pressures (such as antiviral agents or vaccination) may favor replication of some components of a mutant spectrum over others.
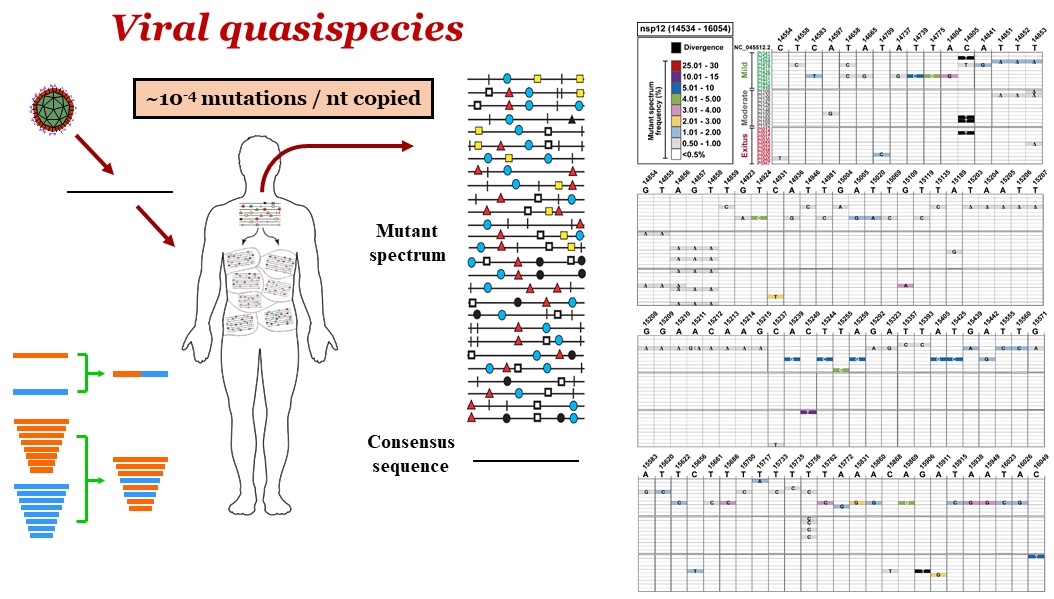
Quasispecies dynamics demands that new approaches be investigated for the prevention and treatment of diseases associated with RNA viruses, to counteract the adaptive capacity conferred by the mutant clouds. The application of ultra-deep sequencing (UDS) methodologies has allowed some penetration into the composition of viral mutant spectra. The main observations on hepatitis C virus (HCV) population dynamics upon extended replication in human hepatoma Huh-7.5 cells are:
- Long-term replication in a non-coevolving cellular environment did not result in HCV population equilibrium. Changes in mutant frequencies (mutational waves) persisted despite increased adaptation to the cell culture environment.
- High HCV fitness confers resistance to non-mutagenic inhibitors, and to the lethal mutagens favipiravir and ribavirin.
- Short term mutational waves were also observed, suggesting that the system develops stochastic perturbations to enhance survival in case of unpredictable change.
- Highly represented substitutions (HRS), different from resistance-associated substitutions (RAS), were identified in virus from patients who failed direct acting antiviral therapies.
- A discrepancy between conserved residues in the Los Alamos database and the mutant spectra of infected patients was observed. This discrepancy could have a negative impact on the efficacy of universal vaccines and pan-genomic antiviral agents.
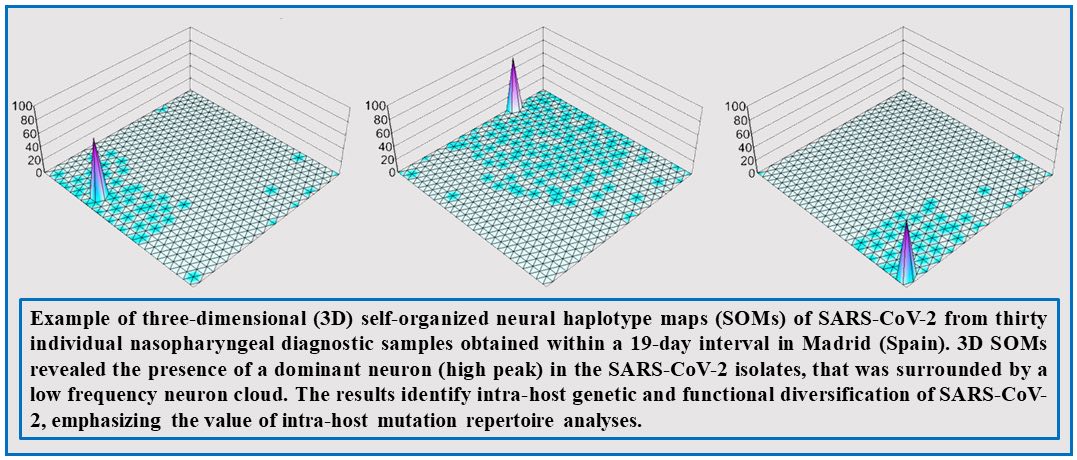
In our laboratory, we want to extend the previous studies on population dynamics to SARS-CoV-2 with the aim of increasing our understanding of quasispecies implications in a comparative manner in cell culture and in vivo. To this aim, we have available more than 8,500 SARS-CoV-2 positive nasopharyngeal swabs from the Fundación Jiménez Díaz in Madrid that cover all pandemic waves. We are currently analyzing the intra-host mutant composition of SARS-CoV-2 populations from infected patients by ultra-deep sequencing, and exploring synergistic combinations between inhibitors to achieve viral extinction. Most of these compounds are nucleotide analogs and some of them act as lethal mutagens, driving virus extinction by an excess of mutations. These projects are performed in collaboration with other teams, as reflected in recent publications.
Publications
Group Members
Group Leader
Celia Perales
Lab assistant
María Eugenia Soria

Funding
News
Un estudio liderado por el CSIC halla una combinación de fármacos eficaz frente al SARS-CoV-2
16 de Abril 2024 La unión de ribavirina y remdesivir consigue eliminar de forma rápida el virus al inducir un exceso de mutaciones en su genoma que le impiden multiplicarse con eficacia El trabajo abre nuevas posibilidades de tratamiento en pacientes vulnerables que...
Identifican mutaciones que afectan la multiplicación del virus SARS-CoV-2 con redes neuronales
27 de Febrero 2024 El estudio ha revelado la aparición incipiente de variantes de coronavirus con propiedades biológicas alteradas en pacientes de COVID-19 Estas mutaciones, hasta ahora inadvertidas por su baja proporción en los pacientes infectados, han sido...