Molecular Virology Of Pathogenic Plus-Strand RNA Viruses
RESEARCH GROUPS

Pablo Gastaminza
Group Leader
Research Summary
Our group focuses on the study of the molecular mechanisms that underlie viral replication and molecular pathogenesis through the study of virus-cell interactions in cell culture models. The main objective of the laboratory is to understand the cellular and molecular mechanisms underlying efficient viral infection, with the ultimate goal of identifying novel diagnostic and therapeutic approaches to fight viral infections and their pathogenesis. The lab uses multidisciplinary approaches studying functional as well as structural aspects of the virus-infected cells. The group has been focused on the hepatitis C virus infection model, but the approaches and methodologies implemented in the laboratory are already being applied to human pathogenic viruses such as dengue, West Nile or SARS-CoV-2.
Research Lines
The laboratory is focused on the study of virus-host interactions through manipulation of the expression of selected host genes by RNAi, CRISPR/Cas9 genome edition or cDNA overexpression and the evaluation of the impact that such manipulation has on different aspects of the viral replication cycle. Our laboratory uses primarily recombinant viruses produced from infectious molecular clones for hepatitis C virus (HCV), dengue virus (DENV) or West Nile virus (WNV). These experimental systems facilitate the study of the function of viral proteins and RNA elements through reverse genetic studies.
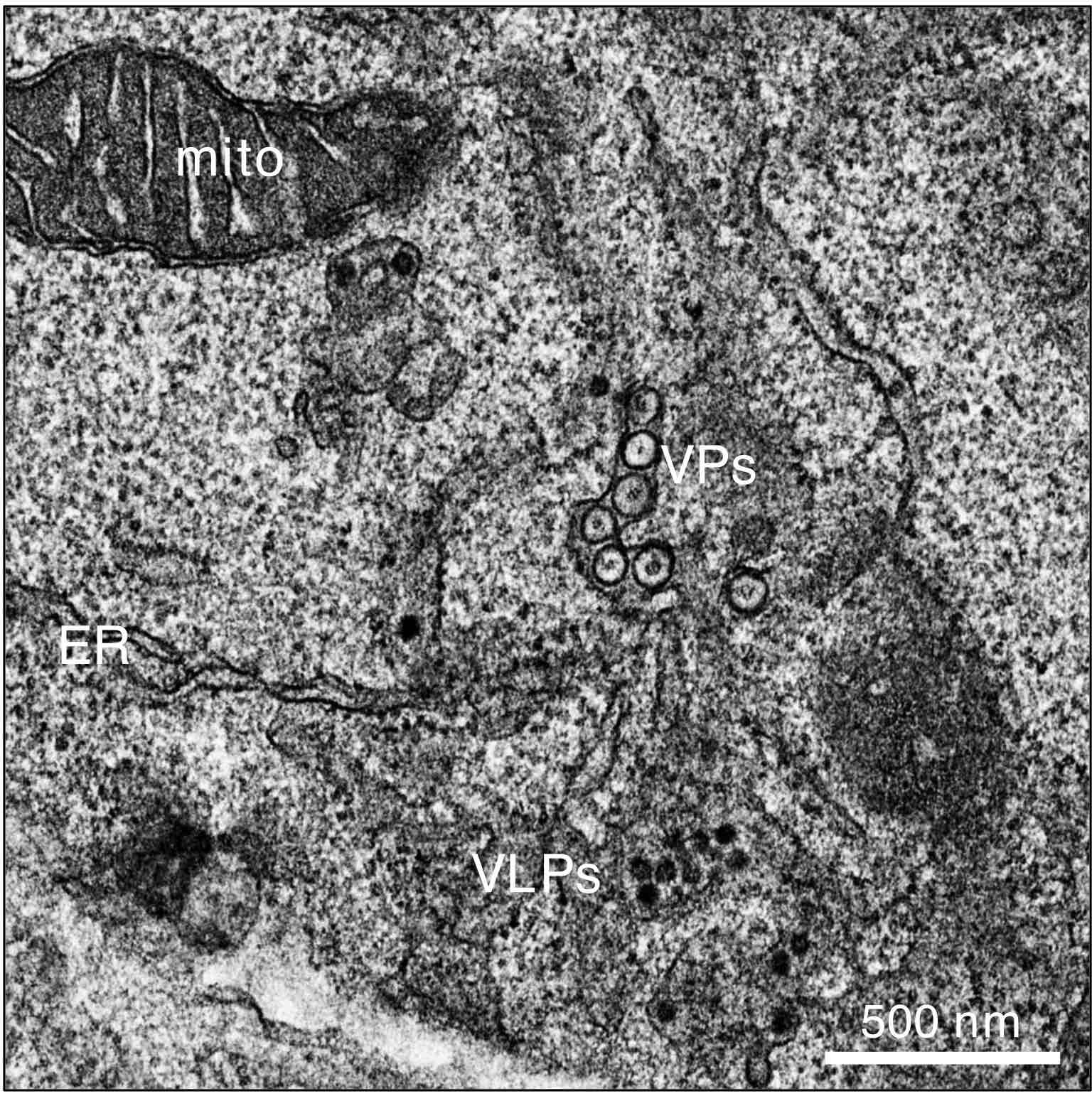
Transmission electron microscopy of cellular and flaviviral structures in West Nile Virus-infected human neuroblasts. Mito, mitochondrion; ER, endoplasmic reticulum; VPs, vesicle packets; VLPs, virus-like particles. Image generated by Victoria Castro, Ph.D. #GastaminzaLab (CNB).
Using these approaches, we were able to show a differential role of two cellular genes involved in glycerophospholipid metabolism (LPIN1 and LPIN2) on the HCV replication cycle (Castro et al 2019; Mingorance et al. 2018). We showed that LPIN1 specifically interferes with early stages of HCV replication at the level replication compartment formation, while LPIN2 interfered with the secretion of infectious virions for different Flaviviridae and other plus-strand RNA viruses. This was due to a strong impact of LPIN2 silencing on the distribution of diacylglycerol, which caused Golgi dispersion and mitochondrial elongation. Similar studies are being carried out on the molecular events underlying the role of the sigma-1 receptor (SIGMAR1) in the establishment of HCV replicase compartments (Friesland et al. 2013). Given that the precise cellular functions of SIGMAR1 are still not completely understood, we have specifically devoted strong efforts in understanding the impact that SIGMAR1 silencing/editing has on host cell functions through integration of unbiased transcriptomic, proteomic and interactomic approaches and implemented “click” chemistry-based approaches to study the fate of host cell proteins under different experimental situations, including after SIGMAR1 silencing.
A second line of research involves the study of the impact of long-term, persistent HCV infection in cell culture on the host cell transcriptome and its reversibility by antiviral treatment. Currently, highly effective direct-acting antiviral (DAA) treatments are available for chronic HCV patients. However, cured HCV patients are still at risk of developing hepatocellular carcinoma, even after elimination of the infection. Thus, we set out to model persistent HCV infection in cell culture using conventional and hepatocyte-like cultures (Castro et al. 2019). Study of the reversion of the transcriptional alterations induced by HCV after antiviral treatment suggest that most, but not all transcriptional alterations are eliminated together with the infection, implying that HCV leaves irreversible “molecular scars” on the host cells. Some of the data obtained in cell culture were also observed in cured patient cohorts, suggesting that some of the abnormal transcriptional traits detected in cell culture could be used in the clinical setup for patient follow up after DAA treatment (Castro et al. 2024).
These functional studies are complemented by the use of different modalities of microscopy to unveil the structural alterations inflicted by viral replication and its reversion by antiviral treatment. In this sense, we have established a long-standing collaboration with the MISTRAL beamline at ALBA synchrotron to produce three-dimensional maps of virus-infected cells in HCV (Pérez-Berná et al. 2016 and 2021), SARS-CoV-2 (Castro et al. 2023) or West Nile viruses (Mamprin et al. in preparation) using soft X-ray tomography, transmission electron microscopy and confocal microscopy. Our group is now integrated in the COCID EU Consortium and is associated with the MSCA CLEXM training network aiming at implementing additional correlative microscopy workflows for the study of viral infection.

Hepatocyte-like cell cultures persistently infected with HCV (top left panel) were treated with DAA until infection was completely eliminated (top right panel) as determined by immunofluorescence microscopy. RNAseq studies were conducted in persistently infected as well as in cured cells and differential transcriptomic profiling is represented with Volcano plots in the absence/presence of DAA treatment (lower panels), suggesting an abnormal transcriptional profile of cured cells.
Publications
Group Members
Group Leader
Pablo Gastaminza
Postdoctoral Researcher
Carlos García-Crespo
Lab Assistant
Gema Calvo Gutierrez
PhD candidates
Emma Diaz Piñero
Victor Venturini Juárez
Sofia Liz Basteiro

Funding
News
Identifican nuevos marcadores para el diagnóstico precoz de patologías hepáticas graves tras la curación de la hepatitis C
18 Julio 2024 Un estudio del CSIC describe genes como marcadores del estado del hígado tras la eliminación del virus de la hepatitis C crónica, que puede dar lugar a enfermedades hepáticas graves como la cirrosis o el carcinoma hepatocelular Aunque los tratamientos...





