Evolution to Tackle Bacterial Infections
Research Project

Sara Hernando-Amado
Project leader
Contact
Research Summary
I study the evolution of mutation-driven antibiotic resistance, with a main interest in the ESKAPE pathogen Pseudomonas aeruginosa.
Research Lines
The main focus of the project is preserving the efficacy of antibiotics. We perform adaptive laboratory evolution experiments to predict and understand the evolution of antibiotic resistance and its associated trade-offs. In particular, we try to understand the evolution of mutation driven-antibiotic resistance, with a main interest in Pseudomonas aeruginosa. This opportunistic pathogen causes nosocomial infections and chronic infections in patients suffering from cystic fibrosis or chronic obstructive pulmonary disease. Its hazard has included this pathogen into the ESKAPE group of bacteria of greatest concern regarding antibiotic resistance.
The use of evolution to curb resistance requires the identification of robust evolutionary trade-offs (as collateral sensitivity, high fitness costs or high metabolic deficiencies) for designing rational therapeutic strategies against bacterial infections using existing antibiotics, something we are focused in. Besides that, we are interested in finding compounds that lead to a transient state of susceptibility (transient collateral sensitivity or synergy) and in identifying physiological weaknesses of antibiotic resistant mutants that can be translated into therapy. Our main objective is not stopping antibiotic resistance but restricting bacterial evolution towards resistance making a rational use of antibiotics, not necessarily new.
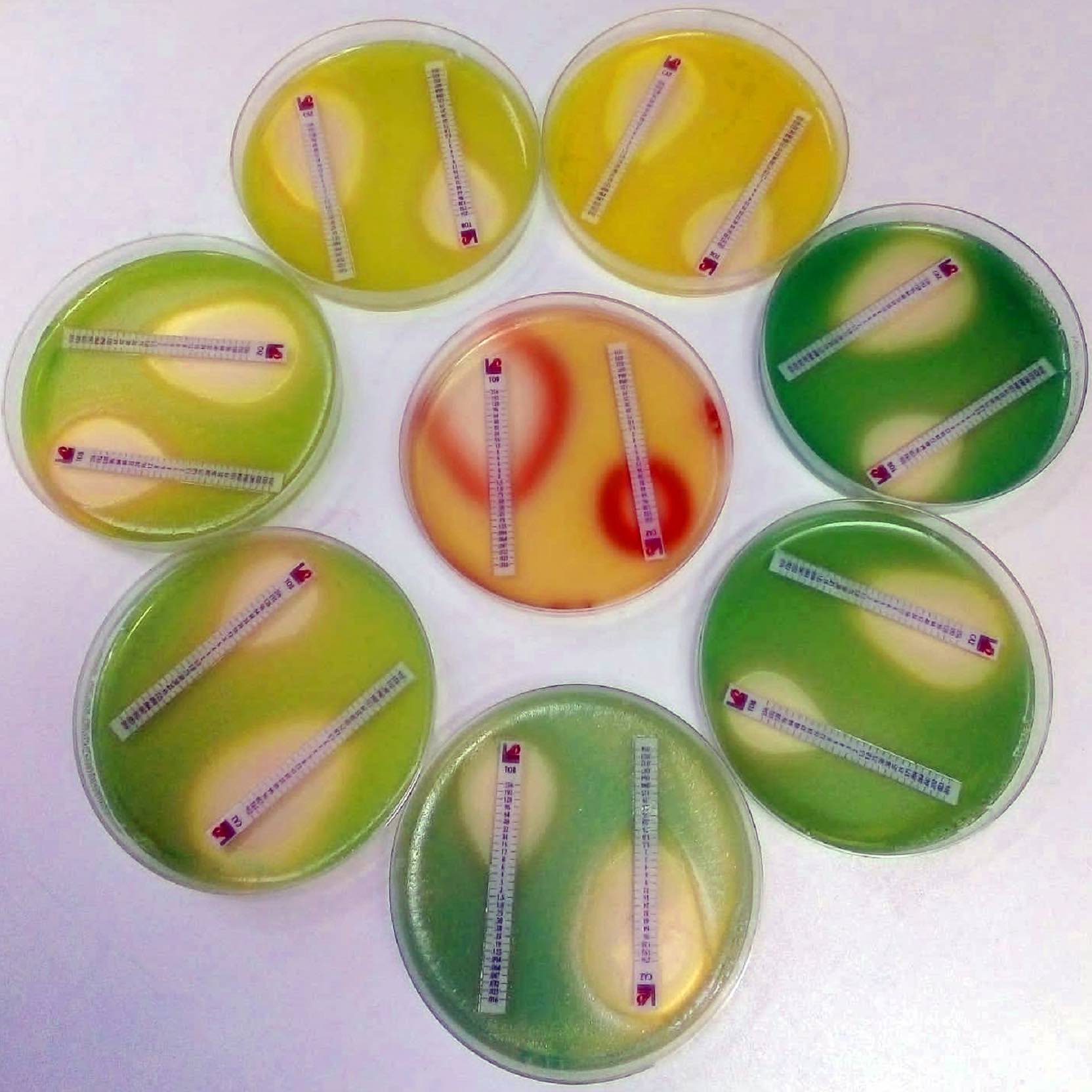
Phenotypic evolutionary landscapes of Pseudomonas aeruginosa in the presence of antibiotics. Adaptive Laboratory Evolution (ALE) experiments allow the prediction of antibiotic resistance evolution in few days, since bacteria rapidly divide. This picture shows antibiotic resistance level (measured with E-Test strips) of the opportunistic pathogen Pseudomonas aeruginosa after just 20 days of evolution.
Publications
Funding

News
Un antibiótico de último recurso selecciona multirresistencia en Pseudomonas aeruginosa
Expertos de 24genetics, el CSIC, Ruber Internacional y Eurofins Megalab debaten sobre el pasado, presente y futuro de la secuenciación genética.



