Regulation of Gene Expression in Plants
RESEARCH GROUP

José Manuel Franco
Group Leader
Research Summary
The main goal in our group is to understand the molecular basis of plant adaptation to changing environmental conditions. In particular, we are focused on the study of the regulation of gene expression as the main regulatory step of gene activity. We are following a combination of experimental and computational approaches for characterization and prediction of the main factors determining the activation of gene expression.
Research Lines
Plant plasticity during adaptation to the environment involves specific transcriptional signal-response networks that allow them to reprogram their growth and development. Regulation of these networks relies on sequence-specific transcription factors (TFs), regulatory proteins responsible for the transcriptional activation or repression of target genes.
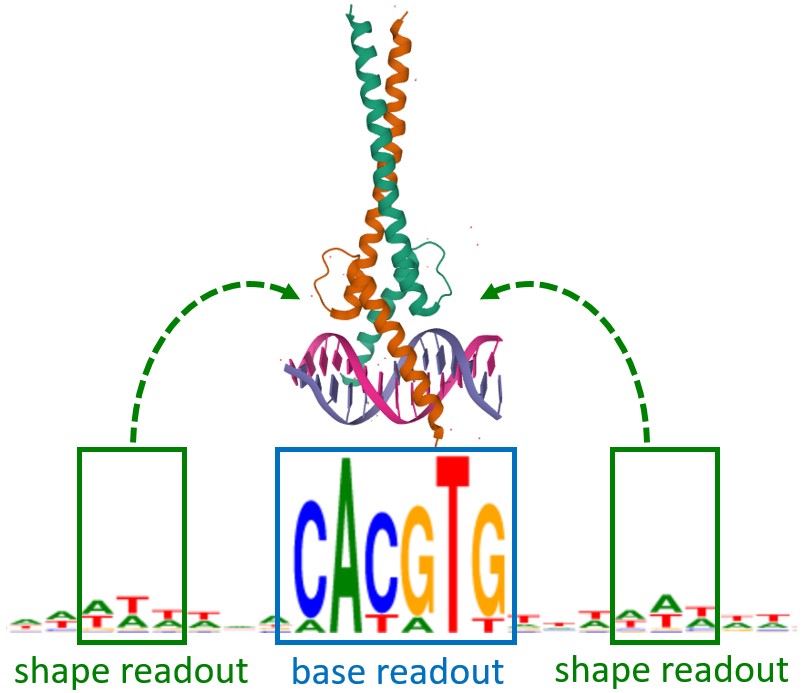
Research in our group is focused in the study of the components that determine specific recognition of TF target genes and which may influence in the levels of gene expression. During the last few years we have contributed to the characterization of one of these components, such as the short DNA sequences bound by TFs, known as TF-binding sites (TFBS). Despite TFBS sequence is the major factor determining target recognition, during the last few years we have explored the role of some other components involved in this process. With this regard, we have demonstrated that binding of some TFs extends beyond the TFBS core sequence, as some distant nucleotides, likely determining DNA-shape, are necessary for protein binding. We are also studying the role of some other components affecting specific recognition of target genes, such the involvement of cytosine methylation in the TFBS region during TF-target recognition, the cooperativity between TFs in DNA-binding and gene activation and the role of intrinsic disordered regions in the TFs during DNA recognition and binding.
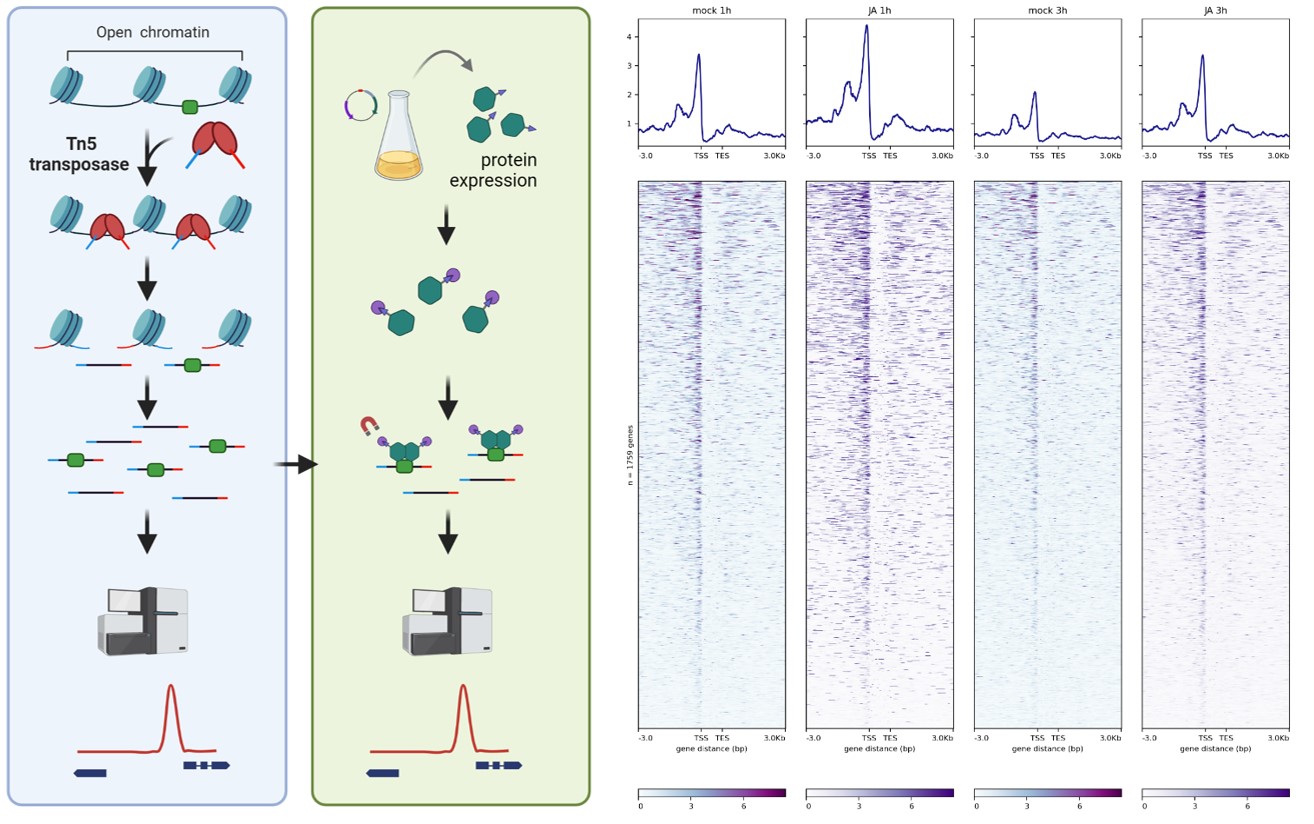
In parallel to the experimental approaches, we are developing some easy-to-use bioinformatic tools useful for the interpretation of transcriptional data and for the prediction of TFBS involved in the regulation of biological processes. These tools would contribute to a better and faster interpretation of biological data for the plant biology community, particularly in the case of non-expert researchers in bioinformatics or in the study of non-model species.
Publications
Group Members
Group Leader
José Manuel Franco
Lab assistant
Marta Godoy Ciudad
PhD candidates
Eva Álvarez Medrano
Joaquín Grau Roldán





