Ecology and Evolution of Bacterial Resistance
RESEARCH GROUPS

José Luis Martínez
Group Leader
Contact
Research Summary
The aim of our work is understanding the mechanisms governing the evolution of antibiotic resistance and the consequences for bacterial physiology of acquiring such resistance. This understanding would serve to implement novel evolution-driven therapeutic approaches to treat more efficiently infections and to reduce antibiotic resistance burden.
Group Members

Research Lines
In the last years, we have standardised some tools, based on experimental evolution, whole-genome sequencing and functional assays, for predicting the evolution of antibiotic resistance and the consequences of acquiring such resistance for bacterial physiology. Using these approaches, we characterised mechanisms of resistance to last-generation antibiotics and combinations of them. Notably, acquisition of resistance may be linked to changes in the susceptibility to other antibiotics, besides those used for selection. In this regard, we have determined the networks of cross-resistance and collateral sensitivity associated to the acquisition of resistance to different antibiotics.
One important focus of our studies is determining the elements that modulate the robustness and predictability of evolutionary trajectories towards antibiotic resistance of bacterial pathogens. Robustness is particularly relevant for exploiting patterns of collateral sensitivity in order to implement more efficient therapeutic strategies based in antibiotic combinations or cycling strategies. We have found some robust collateral sensitivity networks that will be explored in clinical strains in the next future.
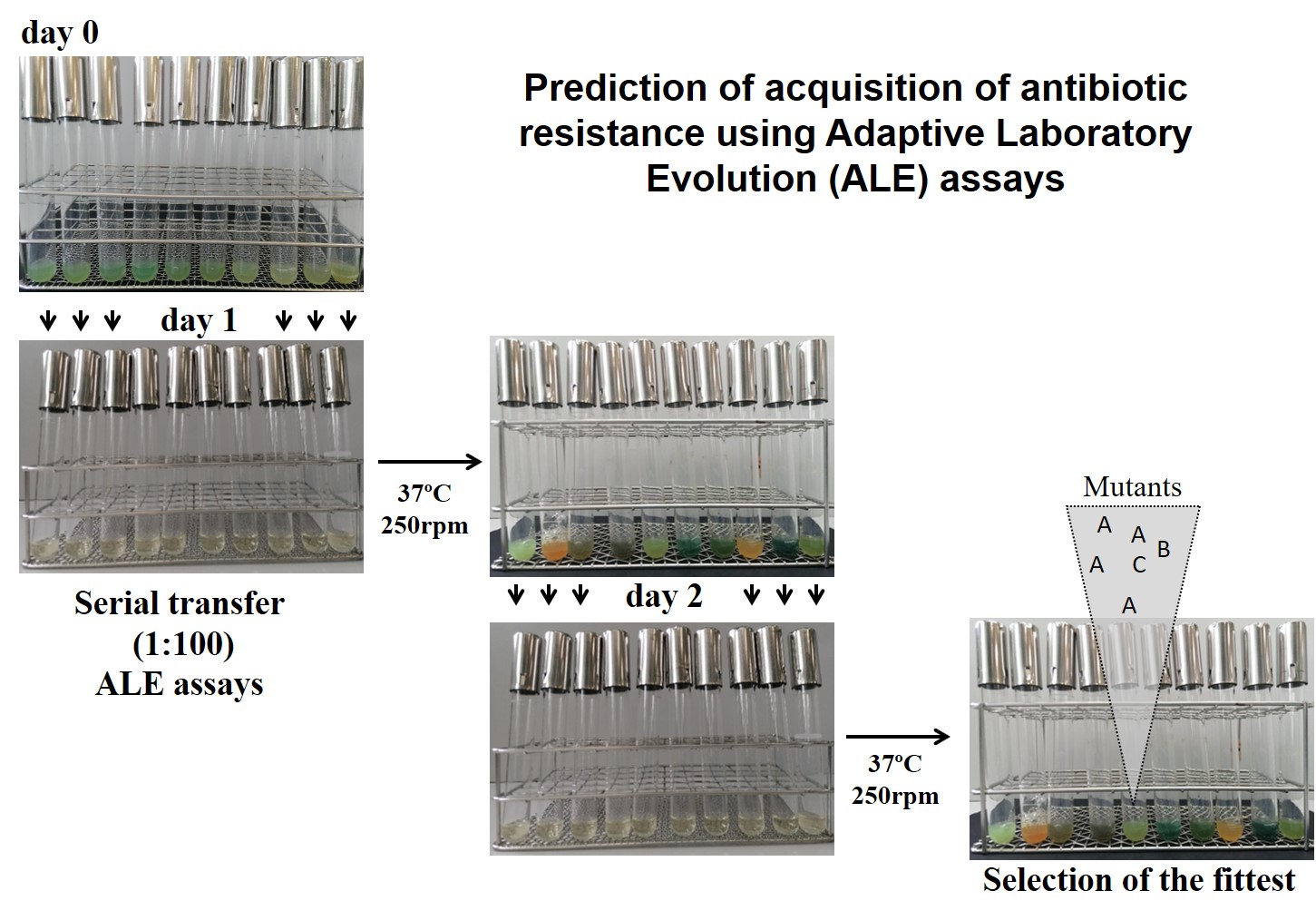
Among the elements that drive the evolution of antibiotic resistance from stochasticity to determinism, we are particularly interested in the epistatic interactions between elements involved in antibiotic resistance and virulence of bacterial pathogens. Besides mutation-driven resistance, we are studying compounds or conditions that can lead to transient resistance, as well as those that can increase the susceptibility to currently used antibiotics and could, hence, be used as co-adjuvants in therapy.
Publications
Laborda, S. Lolle, S. Hernando-Amado, M. Alcalde-Rico, JL. Martínez, S. Molin, H. Krogh Johansen. Mutations in the efflux pump regulator MexZ shift tissue colonization by Pseudomonas aeruginosa to a state of antibiotic tolerance. Nature Communications, (2024) 15, 2584
Hernando-Amado, P. Laborda, JL. Martínez. Tackling antibiotic resistance by inducing transient and robust collateral sensitivity Nature Communications (2023) 14, 1723
Sanz-García, T. Gil-Gil, P. Laborda, P. Blanco, LE Ochoa-Sánchez, F. Baquero, JL Martínez, S. Hernando-Amado. Translating eco-evolutionary biology into therapy to tackle antibiotic resistance Nature Reviews Microbiology (2023), Oct;21(10):671-685.
Gil-Gil, JR Valverde, JL Martínez, F Corona. In vivo genetic analysis of Pseudomonas aeruginosa carbon catabolic repression through the study of CrcZ pseudo-revertants shows that Crc-mediated metabolic robustness is needed for proficient bacterial virulence and antibiotic resistance Microbiology Spectrum (2023), 11:e02350-02323.
S Hernando-Amado, C López-Causapé, P Laborda, F Sanz-García, A Oliver, JL Martínez. Rapid Phenotypic Convergence towards Collateral Sensitivity in Clinical Isolates of Pseudomonas aeruginosa Presenting Different Genomic Backgrounds Microbiology Spectrum (2023), 11:e0227622.
News
Tres investigadores del CNB-CSIC, en la lista de científicos más influyentes del mundo
22 de noviembre 2024 Los investigadores del CNB Luis Enjuanes, José Luis Martínez e Isabel Sola aparecen en la lista Highly Cited Researchers (HCR) del año 2024, elaborada por la plataforma Webofscience Group, de Clarivate Analytics, que reúne a los científicos más...



