Bacterial Engineering for Biomedical Applications
RESEARCH GROUPS

Luis Ángel Fernández
Group Leader
Research Summary
Our research is aimed to engineer E. coli bacteria for biomedical applications, including the selection of small recombinant antibodies and the design of bacteria for diagnostic and therapeutic use in vivo. We study protein secretion systems found in pathogenic E. coli strains and engineer them to develop protein nanomachines that can be applied for selection of recombinant antibodies and the delivery of therapeutic proteins by engineered E. coli strains. Among the recombinant antibodies, we employ single-domain antibodies (sdAbs) or nanobodies, the smallest antibody fragments known-to-date with full antigen-binding capacity. We use synthetic biology approaches and genome engineering to combine the expression of these modular parts in the designed bacteria.
Research Lines
1) Expression and selection of nanobodies against pathogens and cancer. We use E. coli surface display systems to screen libraries of nanobodies for the selection of high-affinity clones that could neutralize viral and bacterial infections. We also select nanobodies binding relevant cell surface antigens associated to cancer to develop targeted therapeutic strategies.
2) Engineering synthetic E. coli bacteria as anti-tumor agents. We use synthetic biology to modify non-pathogenic E. coli chassis with synthetic adhesins and type III protein secretion system (T3SS) to obtain bacteria with specific anti-tumor activities and the ability to deliver specific cargo proteins, including toxins, nanobodies and enzymes.
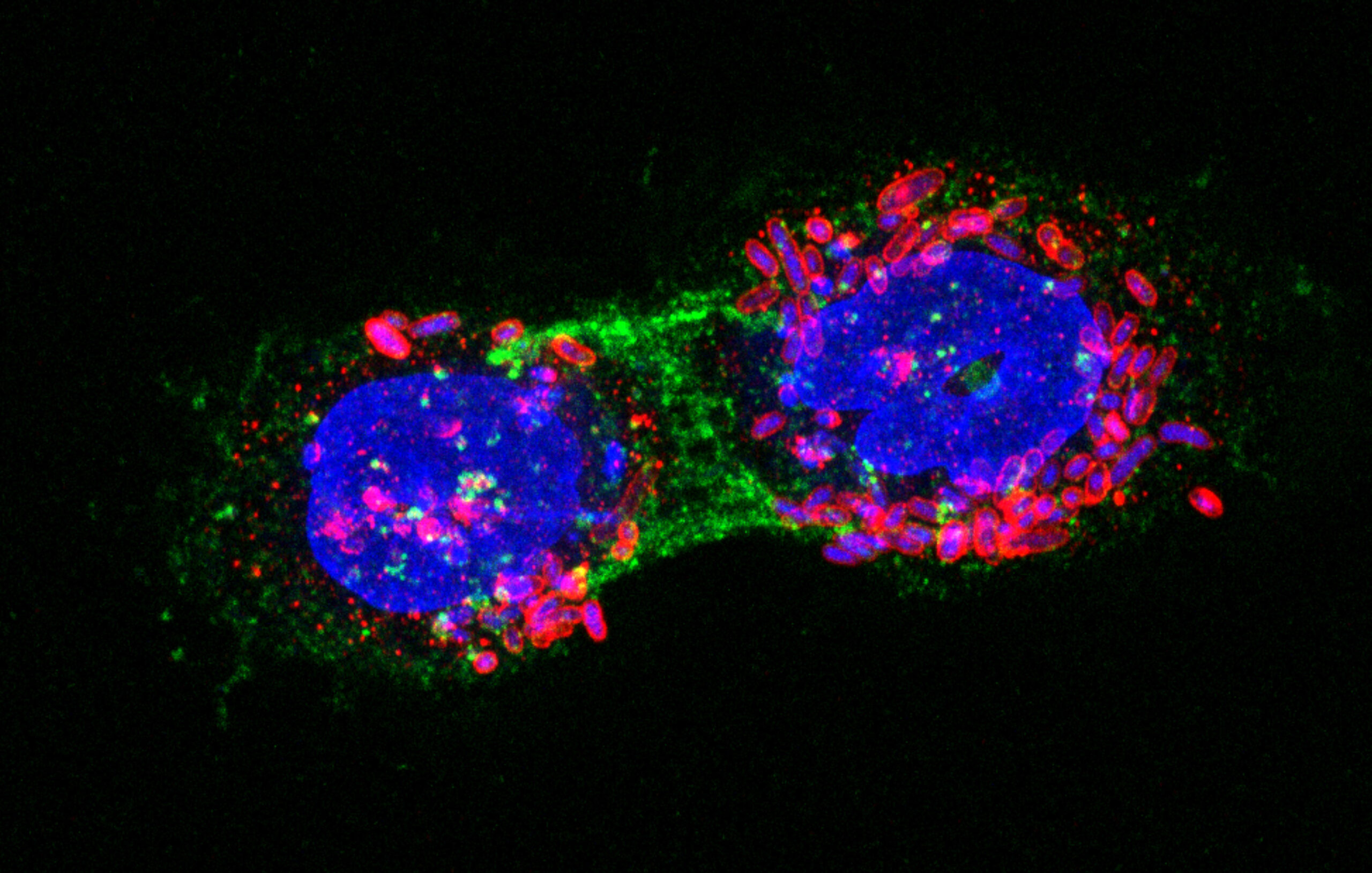
Fluorescence confocal microscopy image showing E. coli bacteria (red fluorescence) with synthetic adhesins targeting an antigen (green fluorescence) expressed on the surface of human tumor cells (nuclei and bacterial DNA stained in blue).
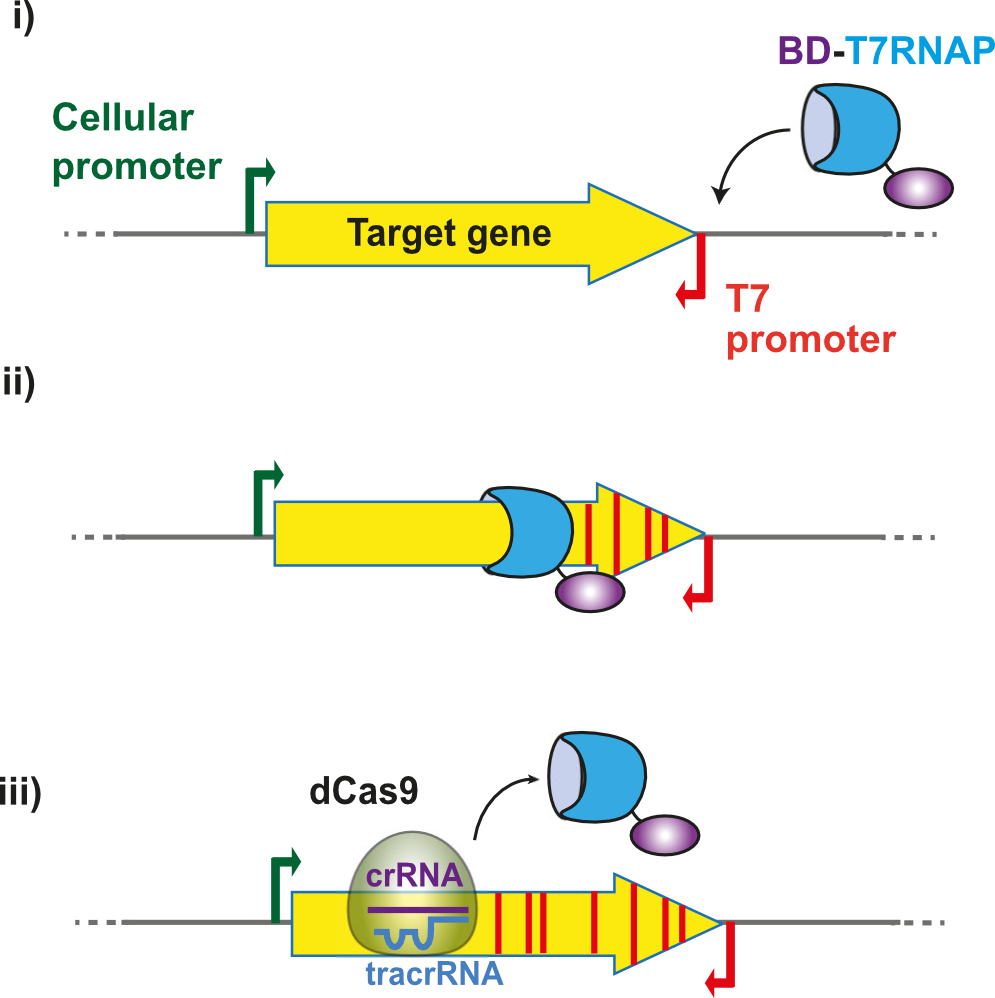
3) Accelerating protein evolution in vivo. We have developed an in vivo mutagenesis system in E. coli, called T7-DIVA, based on the recruitment of base deaminases to a target gene with T7 RNA polymerase. This system enables us to accelerate the directed evolution of proteins of interest, such as enzymes and nanobodies.
Schematic representation of the T7-DIVA in vivo mutagenesis system. The T7 RNA polymerase fusion to a base deaminase (BD-T7RNAP) binds the T7 promoter (i), initiating the transcription and moving along the target gene (yellow filled arrow) introducing mutations (red stripes) in the gene (ii). The fusion stops and detaches from the DNA when encounters a dCas9 molecule bound to a specific sequence determined by the CRISPR RNA (crRNA)(iii).
Publications
Group Members

Group Leader
Luis Ángel Fernández
Lab assistants
Yago Margolles Azpiazu
Mercedes Casanova
Postdoctoral researchers
Lidia Cerdán García
Eva Pico Sánchez
Alejandro Prieto Durán
Staff Scientists
Beatriz Álvarez González
Elena M. Seco Martín
PhD candidates
Ana Ferrández Múrtula
Funding







