Regulation of Gene Expression and Metabolism in Bacteria
RESEARCH GROUPS

Fernando Rojo
Group Leader
Research Summary
Work focuses on the analysis of the global regulatory networks that coordinate and optimize bacterial metabolism. Unravelling the molecular mechanisms involved helps understanding how bacteria coordinate their metabolism and gene expression programs to optimize growth, and aids in the design and optimization of bacterial strains useful in biotechnology. A related research line addresses the role of small RNAs on the regulation of the expression of some of the insertion sequences present in Pseudomonas putida, which is our main model bacterium.
Research Lines
Optimizing the use of a bacterium for a particular biotechnological application requires prior in-depth knowledge of its metabolism, and of the complex regulatory networks that coordinate and optimize the expression of its genome and the configuration of its metabolic fluxes. An additional problem derives from the elements that affect the stability of its genome. Our research addresses these issues in the model bacterium Pseudomonas putida KT2440, which is useful in biotechnology because it is not virulent, it can resist various types of stress, and has a great metabolic versatility.
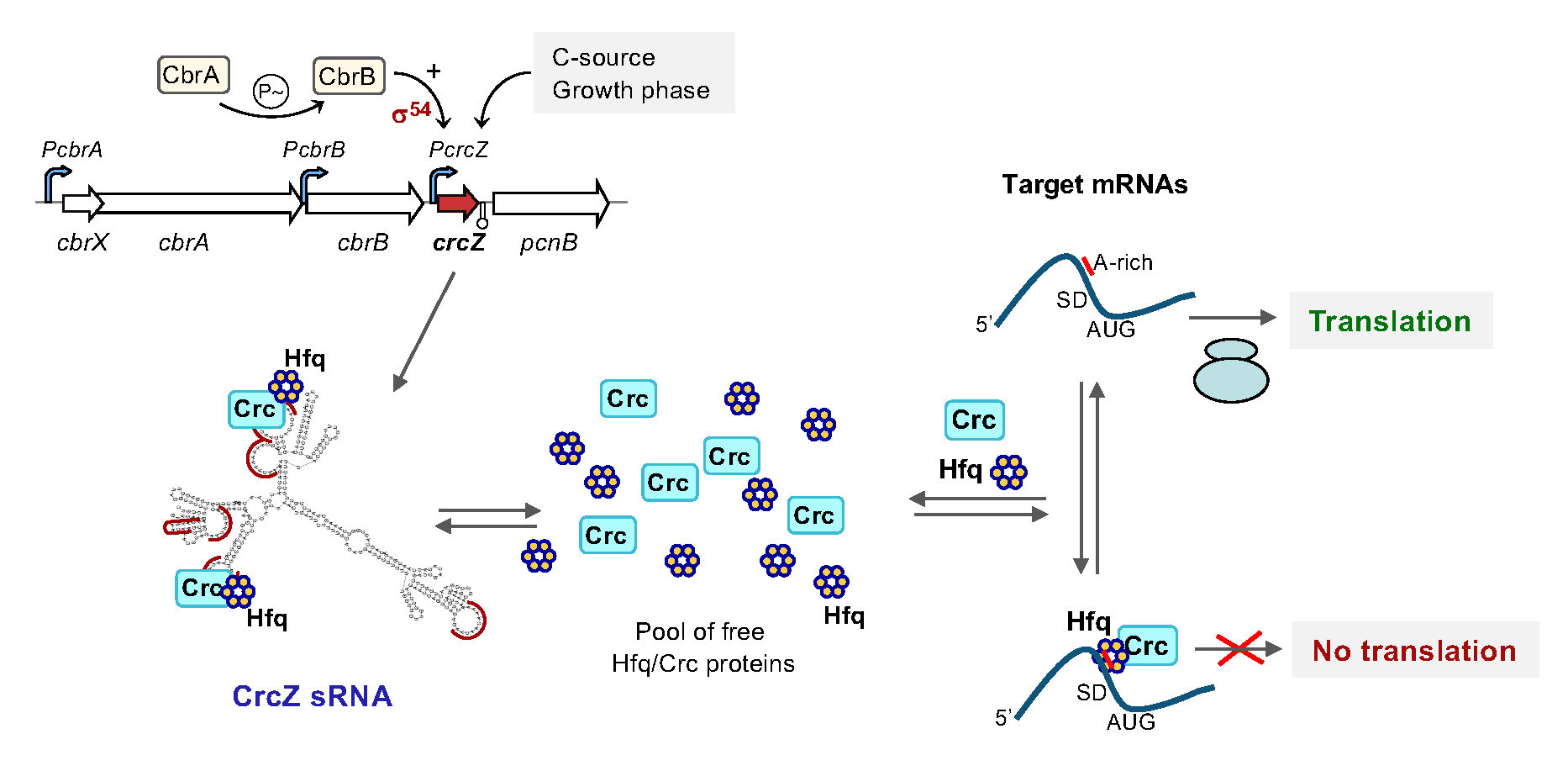
In Pseudomonas, the optimization of metabolic fluxes relies on a regulatory network that includes the Crc and Hfq proteins, and two small RNAs named CrcZ and CrcY. Crc and Hfq inhibit translation of mRNAs containing a specific sequence motif within their translation initiation region, a process that is antagonized by CrcZ and CrcY. We aim to characterise the influence of these regulatory elements on P. putida physiology and the molecular mechanisms involved.
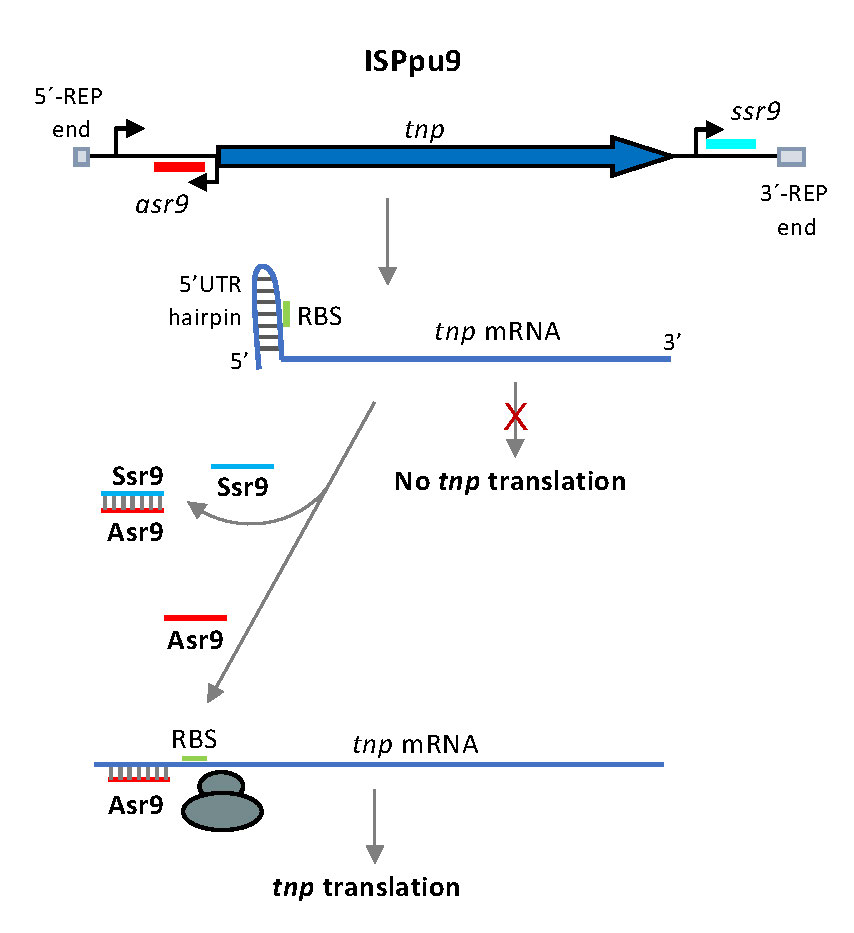
Most bacteria contain diverse mobile genetic elements named Insertion Sequences (IS), which can generate mutations and the inversion or deletion of large DNA segments. Pseudomonas putida KT2440 genome includes 36 insertion sequences, most of them in multiple copies. One of them, named ISPpu9, is present as seven copies inserted at specific intergenic sites named REPs (repetitive intergenic sequences), scattered throughout the chromosome. We are analysing how the activity of this IS is regulated, and how does it mobilise. We have observed that translation of the mRNA encoding the ISPpu9 transposase is inhibited by a highly structured 5’ untranslated region, effect that is counteracted by an antisense small RNA and further modulated by a second small RNA. Current efforts are directed to identify the elements of ISPpu9 involved in its transposition.
Publications
Group Members

Group Leader
Fernando Rojo de Castro
Lab assistant
Luis Yuste
Staff Scientist
Renata Moreno
Postdoctoral Researcher
Elva Quiroz-Rocha
PhD candidates
Bárbara Pérez


