I. Fernández de Castro, R. Tenorio, P. Ortega-González, J.J. Knowlton, P.F. Zamora, C.H. Lee, J.J. Fernández, T.S. Dermody, C. Risco.
Abstract
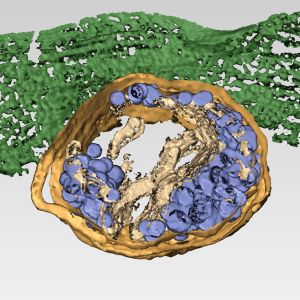
Mammalian orthoreoviruses (reoviruses) are nonenveloped viruses that replicate in cytoplasmic membranous organelles called viral inclusions (VIs) where progeny virions are assembled. To better understand cellular routes of nonlytic reovirus exit, we imaged sites of virus egress in infected, nonpolarized human brain microvascular endothelial cells (HBMECs) and observed one or two distinct egress zones per cell at the basal surface. Transmission electron microscopy and 3D electron tomography (ET) of the egress zones revealed clusters of virions within membrane-bound structures, which we term membranous carriers (MCs), approaching and fusing with the plasma membrane. These virion-containing MCs emerged from larger, LAMP-1-positive membranous organelles that are morphologically compatible with lysosomes. We call these structures sorting organelles (SOs). Reovirus infection induces an increase in the number and size of lysosomes and modifies the pH of these organelles from ∼4.5-5 to ∼6.1 after recruitment to VIs and before incorporation of virions. ET of VI-SO-MC interfaces demonstrated that these compartments are connected by membrane-fusion points, through which mature virions are transported. Collectively, our results show that reovirus uses a previously undescribed, membrane-engaged, nonlytic egress mechanism and highlights a potential new target for therapeutic intervention.
DOI: 10.1083/jcb.201910131.
- The virus sequesters these cellular structures to egress new viral particles
- Cellular factors involved in this trafficking pathway may be key to design new antiviral drugs
Researchers from the National Centre for Biotechnology (CNB-CSIC), have described the mechanism used by human reovirus, a pathogen that causes respiratory and digestive diseases in children and young people, to exit infected cells. This virus, that has also been related to the development of celiac disease, is capable of sequestering cellular lysosomes (organelles attached to the cell membrane) to transport infectious viral particles from inside the cell to the surface. The results of this research that could lead to the design of new antiviral drugs, have been published in the Journal of Cell Biology.
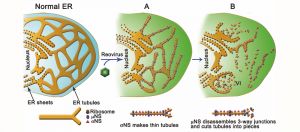
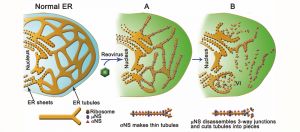
Reoviruses build new organelles (viral factories) in the cytoplasm of infected cells to be able to replicate themselves. This study reveals that the human reovirus modifies and uses endoplasmic reticulum (ER) membranes to produce its replication organelles. The reovirus proteins σNS and μNS are in charge of ER remodeling and the use of this modified ER as scaffold for the assembly of viral factories.
MBio. 2018 Aug 7;9(4). pii: e01253-18. doi: 10.1128/mBio.01253-18.
Tenorio R, Fernández de Castro I, Knowlton JJ, Zamora PF, Lee CH, Mainou BA, Dermody TS, Risco C.
Annu Rev Virol 2014; 1: 453-473.
Risco C, Fernández de Castro I, Sanz-Sánchez L, Narayan K, Grandinetti G, Subramaniam S.
 ReThree-dimensional (3D) imaging technologies are beginning to have significant impact in the field of virology, as they are helping us understand how viruses take control of cells.
ReThree-dimensional (3D) imaging technologies are beginning to have significant impact in the field of virology, as they are helping us understand how viruses take control of cells.
In this article we review several methodologies for 3D imaging of cells and show how these technologies are contributing to the study of viral infections and the characterization of specialized structures formed in virus-infected cells. We include 3D reconstruction by transmission electron microscopy (TEM) using serial sections, electron tomography, and focused ion beam scanning electron microscopy (FIB-SEM).
We summarize from these methods selected contributions to our understanding of viral entry, replication, morphogenesis, egress and propagation, and changes in the spatial architecture of virus-infected cells. In combination with live-cell imaging, correlative microscopy, and new techniques for molecular mapping in situ, the availability of these methods for 3D imaging is expected to provide deeper insights into understanding the structural and dynamic aspects of viral infection.
Los virus ARN, entre los que se encuentran muchos patógenos para humanos, animales y plantas, replican sus genomas en membranas del interior de las células. En estas membranas los virus reclutan factores celulares que participan en el ensamblaje y actividades de los complejos replicativos. Estos agregados macromoleculares, que suelen ensamblarse en vesículas con aperturas al citosol o “esférulas”, se encargan de fabricar múltiples copias del genoma viral que se incorporarán posteriormente en las nuevas partículas virales infectivas. Aunque se han identificado numerosos factores necesarios para el ensamblaje funcional de los orgánulos de replicación viral se desconoce la función exacta de la mayoría de ellos.
 En colaboración con investigadores de la Universidad de Kentucky en los Estados Unidos, el laboratorio del Centro Nacional de Biotecnología del CSIC (CNB) dirigido por Cristina Risco han encontrado evidencias de una función inesperada y sorprendente que desempeña la proteína celular Vps4 en la replicación de un virus ARN perteneciente a la familia de los Tombusvirus, patógenos que infectan plantas y que causan importantes pérdidas en cosechas.
En colaboración con investigadores de la Universidad de Kentucky en los Estados Unidos, el laboratorio del Centro Nacional de Biotecnología del CSIC (CNB) dirigido por Cristina Risco han encontrado evidencias de una función inesperada y sorprendente que desempeña la proteína celular Vps4 en la replicación de un virus ARN perteneciente a la familia de los Tombusvirus, patógenos que infectan plantas y que causan importantes pérdidas en cosechas.
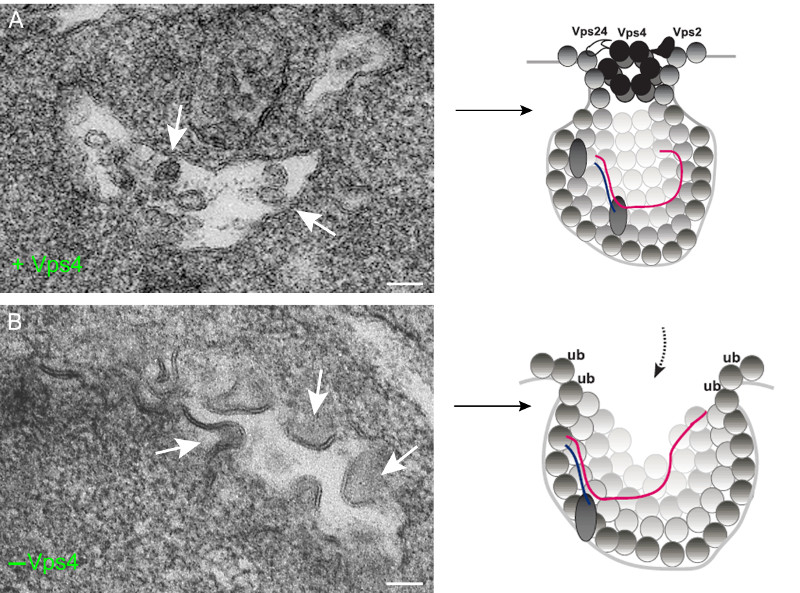
Vps4 es un componente de las proteínas celulares ESCRT y de la familia de las ATPasas AAA+ que usan ATP para remodelar estructuras macromoleculares en las células. Vps4 participa en una variedad de procesos biológicos como por ejemplo la fusión de membranas para la formación de vesículas. Utilizando nuevas técnicas de imagen en microscopía electrónica y según acaban de publicar en la revista PLoS Pathogens, los autores han demostrado que Vps4 participa en la estabilización del poro o cuello de las esférulas a través del cual se produce el intercambio de materiales con el citosol. Esta apertura controlada es necesaria para que el complejo replicativo desempeñe sus funciones y para que el ARN viral quede protegido de la degradación por nucleasas.
A diferencia de lo que ocurre en los procesos celulares habituales en los que participa, Vps4 se incorpora de manera estable en las vesículas virales o esférulas y pasa a formar parte del complejo replicativo. De hecho cuando Vps4 no está presente se forman esférulas totalmente abiertas, sin constricción o cuello y en las que el ARN viral de nueva síntesis está desprotegido. Los autores proponen que Vps4 y otras proteínas ESCRT son indispensables para la deformación de las membranas y el ensamblaje de los complejos replicativos de Tombusvirus. Es muy probable que otros virus de plantas y animales que transforman membranas celulares para construir esférulas también utilicen estas proteínas para ensamblar sus replicasas y proteger sus genomas recién sintetizados.
- Barajas D, Martín IFdC, Pogany J, Risco C, Nagy PD. Noncanonical role for the host Vps4 AAA+ ATPase ESCRT protein in the formation of Tomato bushy stunt virus replicase. PLoS Pathog. 2014; 10(4): e1004087.
PLoS Pathog 2014; 10(4): e1004087.
Barajas D, Martín IFdC, Pogany J, Risco C, Nagy PD.
Assembling of the membrane-bound viral replicase complexes (VRCs) consisting of viral- and host-encoded proteins is a key step during the replication of positive-stranded RNA viruses in the infected cells. Previous genome-wide screens with Tomato bushy stunt tombusvirus (TBSV) in a yeast model host have revealed the involvement of eleven cellular ESCRT (endosomal sorting complexes required for transport) proteins in viral replication. The ESCRT proteins are involved in endosomal sorting of cellular membrane proteins by forming multiprotein complexes, deforming membranes away from the cytosol and, ultimately, pinching off vesicles into the lumen of the endosomes.
In this paper, we show an unexpected key role for the conserved Vps4p AAA+ ATPase, whose canonical function is to disassemble the ESCRT complexes and recycle them from the membranes back to the cytosol. We find that the tombusvirus p33 replication protein interacts with Vps4p and three ESCRT-III proteins. Interestingly, Vps4p is recruited to become a permanent component of the VRCs as shown by co-purification assays and immuno-EM. Vps4p is co-localized with the viral dsRNA and contacts the viral (+)RNA in the intracellular membrane. Deletion of Vps4p in yeast leads to the formation of crescent-like membrane structures instead of the characteristic spherule and vesicle-like structures. The in vitro assembled tombusvirus replicase based on cell-free extracts (CFE) from vps4Δ yeast is highly nuclease sensitive, in contrast with the nuclease insensitive replicase in wt CFE.
These data suggest that the role of Vps4p and the ESCRT machinery is to aid building the membrane-bound VRCs, which become nuclease-insensitive to avoid the recognition by the host antiviral surveillance system and the destruction of the viral RNA. Other (+)RNA viruses of plants and animals might also subvert Vps4p and the ESCRT machinery for formation of VRCs, which require membrane deformation and spherule formation.
MBio. 2014; 5(1). pii: e00931-13.
Fernández de Castro I, Zamora PF, Ooms L, Fernández JJ, Lai CM, Mainou BA, Dermody TS, Risco C.
 Most viruses that replicate in the cytoplasm of host cells form neo-organelles that serve as sites of viral genome replication and particle assembly. These highly specialized structures concentrate viral replication proteins and nucleic acids, prevent the activation of cell-intrinsic defenses, and coordinate the release of progeny particles. Despite the importance of inclusion complexes in viral replication, there are key gaps in the knowledge of how these organelles form and mediate their functions.
Most viruses that replicate in the cytoplasm of host cells form neo-organelles that serve as sites of viral genome replication and particle assembly. These highly specialized structures concentrate viral replication proteins and nucleic acids, prevent the activation of cell-intrinsic defenses, and coordinate the release of progeny particles. Despite the importance of inclusion complexes in viral replication, there are key gaps in the knowledge of how these organelles form and mediate their functions.
Reoviruses are nonenveloped, double-stranded RNA (dsRNA) viruses that serve as tractable experimental models for studies of dsRNA virus replication and pathogenesis. Following reovirus entry into cells, replication occurs in large cytoplasmic structures termed inclusions that fill with progeny virions. Reovirus inclusions are nucleated by viral nonstructural proteins, which in turn recruit viral structural proteins for genome replication and particle assembly. Components of reovirus inclusions are poorly understood, but these structures are generally thought to be devoid of membranes. We used transmission electron microscopy and three-dimensional image reconstructions to visualize reovirus inclusions in infected cells.
These studies revealed that reovirus inclusions form within a membranous network. Viral inclusions contain filled and empty viral particles and microtubules and appose mitochondria and rough endoplasmic reticulum (RER). Immunofluorescence confocal microscopy analysis demonstrated that markers of the ER and ER-Golgi intermediate compartment (ERGIC) codistribute with inclusions during infection, as does dsRNA. dsRNA colocalizes with the viral protein σNS and an ERGIC marker inside inclusions. These findings suggest that cell membranes within reovirus inclusions form a scaffold to coordinate viral replication and assembly.
COOKIES POLICY
A cookie is a text file that is stored on your computer or mobile device via a web server and only that server will be able to retrieve or read the contents of the cookie and allow the Web site remember browser preferences and navigate efficiently. Cookies make the interaction between the user and the website faster and easier.
General information
This Website uses cookies. Cookies are small text files generated by the web pages you visit, which contain the session data that can be useful later in the website. In this way this Web remembers information about your visit, which can facilitate your next visit and make the website more useful.
How do cookies?
Cookies can only store text, usually always anonymous and encrypted. No personal information is ever stored in a cookie, or can be associated with identified or identifiable person.
The data allow this website to keep your information between the pages, and also to discuss how to interact with the website. Cookies are safe because they can only store information that is put there by the browser, which is information the user entered in the browser or included in the page request. You can not run the code and can not be used to access your computer. If a website encrypts cookie data, only the website can read the information.
What types of cookies used?
The cookies used by this website can be distinguished by the following criteria:
1. Types of cookies as the entity that manages:
Depending on who the entity operating the computer or domain where cookies are sent and treat the data obtained, we can distinguish:
- Own cookies: are those that are sent to the user's terminal equipment from a computer or domain managed by the editor itself and from which provides the service requested by the user.
- Third party cookies: these are those that are sent to the user's terminal equipment from a machine or domain that is not managed by the publisher, but by another entity data is obtained through cookies.
In the event that the cookies are installed from a computer or domain managed by the editor itself but the information collected by these is managed by a third party can not be considered as party cookies.
2. Types of cookies as the length of time that remain active:
Depending on the length of time that remain active in the terminal equipment can be distinguished:
- Session cookies: cookies are a type designed to collect and store data while the user accesses a web page. Are usually used to store information that only worth preserving for the service requested by the user at any one time (eg a list of products purchased).
- Persistent cookies: cookies are a type of data which are stored in the terminal and can be accessed and treated for a period defined by the head of the cookie, and can range from a few minutes to several years.
3. Cookies types according to their purpose:
Depending on the purpose for which the data are processed through cookies, we can distinguish between:
- Technical cookies: these are those that allow the user to navigate through a web page or application platform and the use of different options or services it exist as, for example, control traffic and data communication, identify the session, access to restricted access parts, remember the elements of an order, make the buying process an order, make an application for registration or participation in an event, use security features while browsing store content for dissemination videos or sound or share content via social networks.
- Customization cookies: these are those that allow the user to access the service with some general characteristics based on a predefined set of criteria in the user terminal would eg language, the type of browser through which you access the service, the locale from which you access the service, etc.
- Analysis cookies: they are those that allow the responsible for them, monitoring and analyzing the behavior of users of the web sites that are linked. The information gathered through such cookies are used in measuring the activity of web sites, application or platform and for the profiling of user navigation of such sites, applications and platforms, in order to make improvements function data analysis how users use the service.
Management tool cookies
This Website uses Google Analytics.
Google Analytics is a free tool from Google that primarily allows website owners know how users interact with your website. Also, enable cookies in the domain of the site in which you are and uses a set of cookies called "__utma" and "__utmz" to collect information anonymously and reporting of website trends without identifying individual users..
For statistics of use of this website use cookies in order to know the level of recurrence of our visitors and more interesting content. This way we can concentrate our efforts on improving the most visited areas and make the user more easily find what they are looking for. On this site you can use the information from your visit for statistical evaluations and calculations anonymous data and to ensure the continuity of service or to make improvements to their websites. For more details, see the link below privacy policy [http://www.google.com/intl/en/policies/privacy/]
How to manage cookies on your computer: disabling and deleting cookies
All Internet browsers allow you to limit the behavior of a cookie or disable cookies within settings or browser settings. The steps for doing so are different for each browser, you can find instructions in the help menu of your browser.
If you decline the use of cookies, since it is possible thanks to the preferences menu of your browser or settings, reject, this website will continue to function properly without the use of the same.
Can you allow, block or delete cookies installed on your computer by setting your browser options installed on your computer:
- For more information about Internet Explorer click here.
- For more information on Chrome click here.
- For more information about Safari click here.
- For more information about Firefox click here.
Through your browser, you can also view the cookies that are on your computer, and delete them as you see fit. Cookies are text files, you can open and read the contents. The data within them is almost always encrypted with a numeric key corresponding to an Internet session so often has no meaning beyond the website who wrote it.
Informed consent
The use of this website on the other hand, implies that you paid your specific consent to the use of cookies, on the terms and conditions provided in this Cookies Policy, without prejudice to the measures of deactivation and removal of cookies that you can take, and mentioned in the previous section.